Microarray Evaluation of Antimicrobial Resistance and Virulence of Escherichia coli Isolates from Portuguese Poultry
Abstract
:1. Introduction
2. Results
| Antibiotic | Type of Poultry Production | Total (n = 174) | |||
|---|---|---|---|---|---|
| Intensive Meat Production Farms (n = 121) | Extensive Meat Production Farms (n = 11) | Intensive Layers Farms (n = 17) | Intensive Breeding Farms (n = 25) | ||
| Tetracycline | 90 (74%) | 8 (73%) | 10 (59%) | 13 (52%) | 121 (70%) |
| Ampicillin | 80 (66%) | 7 (64%) | 7 (41%) | 15 (60%) | 109 (63%) |
| Mezlocillin | 77 (64%) | 6 (55%) | 7 (41%) | 12 (48%) | 102 (59%) |
| Piperacillin | 73 (60%) | 5 (45%) | 7 (41%) | 12 (48%) | 97 (56%) |
| Moxifloxacin | 73 (60%) | 6 (55%) | 9 (53%) | 10 (40%) | 98 (56%) |
| Ciprofloxacin | 65 (54%) | 5 (45%) | 8 (47%) | 8 (32%) | 86 (49%) |
| Norfloxacin | 66 (55%) | 5 (45%) | 6 (35%) | 8 (32%) | 85 (49%) |
| Cefazolin | 43 (36%) | 5 (45%) | 1 (6%) | 9 (36%) | 58 (33%) |
| Trimethoprim-sulfamethoxazole | 46 (38%) | 5 (45%) | 1 (6%) | 5 (20%) | 57 (33%) |
| Cefpodoxime | 42 (35%) | 5 (45%) | 0 (0%) | 8 (32%) | 55 (32%) |
| Ampicillin plus sulbactam | 34 (28%) | 4 (36%) | 2 (12%) | 4 (16%) | 44 (25%) |
| Levofloxacin | 37 (31%) | 3 (27%) | 1 (6%) | 2 (8%) | 43 (25%) |
| Cefotaxime | 26 (21%) | 3 (27%) | 1 (6%) | 7 (28%) | 37 (21%) |
| Cefuroxime | 23 (19%) | 2 (18%) | 1 (6%) | 5 (20%) | 31 (18%) |
| Amoxicillin plus clavulanic acid | 22 (18%) | 3 (27%) | 0 (0%) | 4 (16%) | 29 (17%) |
| Gentamicin | 20 (17%) | 0 (0%) | 3 (18%) | 6 (24%) | 29 (17%) |
| Aztreonam | 19 (16%) | 1 (9%) | 0 (0%) | 6 (24%) | 26 (15%) |
| Cefoxitin | 19 (16%) | 2 (18%) | 0 (0%) | 3 (12%) | 24 (14%) |
| Chloramphenicol | 13 (11%) | 2 (18%) | 2 (12%) | 4 (16%) | 21 (12%) |
| Ceftazidime | 12 (10%) | 0 (0%) | 0 (0%) | 7 (28%) | 19 (11%) |
| Tobramycin | 14 (12%) | 0 (0%) | 1 (6%) | 3 (12%) | 18 (10%) |
| Cefepime | 9 (7%) | 2 (18%) | 0 (0%) | 2 (8%) | 13 (7%) |
| Colistin | 7 (6%) | 0 (0%) | 0 (0%) | 5 (20%) | 12 (7%) |
| Nitrofurantoin | 6 (5%) | 0 (0%) | 1 (6%) | 0 (0%) | 7 (4%) |
| Piperacillin plus tazobactam | 1 (1%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1%) |
| Fosfomycin | 1 (1%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1%) |
| Ertapenem | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Imipenem | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Meropenem | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Amikacin | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Tigecycline | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
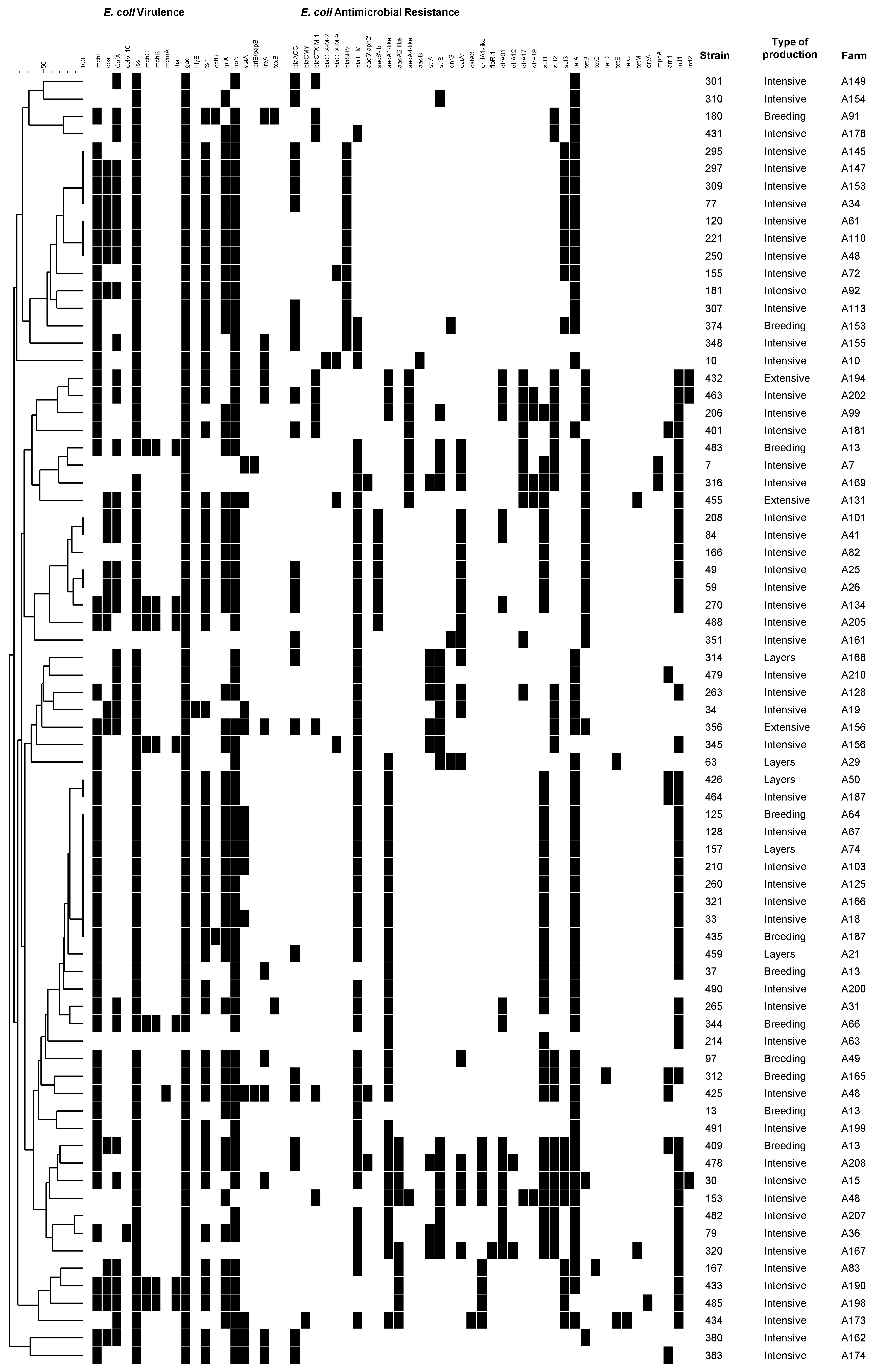
3. Discussion
4. Experimental Section
4.1. Bacterial Isolation and Phenotypic Resistance Typing
4.2. Microarray Analyses and Data Analysis
4.3. Statistical Analysis
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- European Medicines Agency (EMEA). Public Statement on the Use of (Fluoro) Quinolones in Food-Producing Animals in the EUROPEAN Union: Development of Resistance and Impact on the Human and Animal Health; EMEA/CVMP/SAGAM/184651/2005; EMEA: London, UK, 2007. [Google Scholar]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2011. EFSA J. 2013, 11. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2012. EFSA J. 2014, 12. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC). Antimicrobial Resistance Surveillance in Europe 2012. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net); ECDC: Stockholm, Sweden, 2013; p. 218. [Google Scholar]
- Hölzel, C.S.; Harms, K.S.; Bauer, J.; Bauer-Unkauf, I.; Hörmansdorfer, S.; Kämpf, P.; Mölle, G.; Oehme, C.; Preikschat, P.; Schwaiger, K. Diversity of antimicrobial resistance genes and class-1-integrons in phylogenetically related porcine and human Escherichia coli. Vet. Microbiol. 2012, 160, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Kariyawasam, S.; Wannemuehler, Y.; Mangiamele, P.; Johnson, S.J.; Doetkott, C.; Skyberg, J.A.; Lynne, A.M.; Johnson, J.R.; Nolan, L.K. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 2007, 189, 3228–3236. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, L.; Liu, C.M.; Price, L.B. Foodborne urinary tract infections: A new paradigm for antimicrobial-resistant foodborne illness. Front. Microbiol. 2013, 4, e29. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; Paterson, D.L.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Garau, J. Other antimicrobials of interest in the era of extended-spectrum β-lactamases: Fosfomycin, nitrofurantoin and tigecycline. Clin. Microbiol. Infect. 2008, 14, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Calhau, V.; Domingues, S.; Ribeiro, G.; Mendonça, N.; da Silva, G.J. Interplay between pathogenicity island carriage, resistance profile and plasmid acquisition in uropathogenic Escherichia coli. J. Med. Microbiol. 2015, 64, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Card, R.; Zhang, J.; Das, P.; Cook, C.; Woodford, N.; Anjum, M.F. Evaluation of an expanded microarray for detecting antibiotic resistance genes in a broad range of Gram-negative bacterial pathogens. Antimicrob. Agents Chemother. 2013, 57, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, N.; Leitão, J.; Manageiro, V.; Ferreira, E.; ARSIP; Caniça, M. Spread of clinical extended-spectrum ß-lactamase CTX-M-producing Escherichia coli isolates in community and nosocomial environments in Portugal. Antimicrob. Agents Chemother. 2007, 51, 1946–1955. [Google Scholar]
- Zhang, T.; Wang, C.G.; Lv, J.C.; Wang, R.S.; Zhong, X.H. Survey on tetracycline resistance and antibiotic-resistant genotype of avian Escherichia coli in North China. Poult. Sci. 2012, 91, 2774–2777. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Gomes, A.; Lino, C.M.; Pena, A. Does the use of fluoroquinolones in poultry production reach your plate? Portuguese nationwide surveillance. In Proceedings of the 8th Conference on Sustainable Development of Energy Water and Environment Systems (ISSN 1847-7178), Dubrovnik, Hrvatska, 22–27 September 2013; Faculty of Mechanical Engineering and Naval Architecture: Dubrovnik, Croatia, 2013. [Google Scholar]
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.M.; Nolan, L.K. Evolution of the iss gene in Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2360–2369. [Google Scholar] [CrossRef] [PubMed]
- Anjum, M.F.; Mafura, M.; Slickers, P.; Ballmer, K.; Kuhnert, P.; Woodward, M.J.; Ehricht, R. Pathotyping Escherichia coli by using miniaturized DNA microarrays. Appl. Environ. Microbiol. 2007, 73, 5692–5697. [Google Scholar] [CrossRef] [PubMed]
- Welch, R.A.; Burland, V.; Plunkett III, G.; Redford, P.; Roesch, P.; Rasko, D.; Buckles, E.L.; Liou, S.-R.; Boutin, A.; Hackett, J.; et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 17020–17024. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 1991, 4, 80–128. [Google Scholar] [PubMed]
- Heimer, S.R.; Rasko, D.A.; Lockatell, C.V.; Johnson, D.E.; Mobley, H.L. Autotransporter genes pic and tsh are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infect. Immun. 2004, 72, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Milflores-Flores, L.; Garcia-Gallegos, J.G.; Patel, S.D.; Best, A.; la Ragione, R.M.; Martinez-Laguna, Y.; Woodward, M.J. Environmental regulation and colonization attributes of the long polar fimbriae (LPF) of Escherichia coli O157:H7. Int. J. Med. Microbiol. 2007, 297, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Maturana, V.G.; de Pace, F.; Carlos, C.; Mistretta Pires, M.; Amabile-de-Campos, T.; Nakazato, G.; Guedes-Stheling, E.; Logue, C.M.; Nolan, L.K.; Dias-da-Silveira, W. Subpathotypes of Avian Pathogenic Escherichia coli (APEC) Exist as Defined by their Syndromes and Virulence Traits. Open Microbiol. J. 2011, 5, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Jelacic, S.; Schoening, L.M.; Clabots, C.; Shaikh, N.; Mobley, H.L.; Tarr, P.I. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect. Immun. 2005, 73, 965–971. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, E.; Gaggero, C.; Laviña, M. The structural gene for Microcin H47 encodes a peptide precursor with antibiotic activity. Antimicrob. Agents Chemother. 1999, 43, 2176–2182. [Google Scholar] [PubMed]
- International Organization for Standardization. Microbiology of Food and Animal Feeding Stuffs. Horizontal Method for the Detection and Enumeration of Presumptive Escherichia coli. Most Probable Number Technique; ISO 7251:2005; International Organization for Standardization IOS: Geneva, Switzerland, 2005; p. 13. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement M100-S20; CLSI: Wayne, PA, USA, 2010; p. 188. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 4.0. 2014. Available online: http://www.eucast.org (accessed on 7 January 2016).
- Figueiredo, R.; Card, R.; Nunes, C.; AbuOun, M.; Bagnall, M.C.; Nunez, J.; Anjum, M.F.; da Silva, G.J. Virulence Characterization of Salmonella enterica by a New Microarray: Detection and Evaluation of the Cytolethal Distending Toxin Gene Activity in the Unusual Host S. Typhimurium. PLoS ONE 2015, 10, e0135010. [Google Scholar] [CrossRef] [PubMed]
- Szmolka, A.; Anjum, M.F.; la Ragione, R.M.; Kaszanyitzky, E.J.; Nagy, B. Microarray based comparative genotyping of gentamicin resistant Escherichia coli strains from food animals and humans. Vet. Microbiol. 2012, 156, 110–118. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendonça, N.; Figueiredo, R.; Mendes, C.; Card, R.M.; Anjum, M.F.; Da Silva, G.J. Microarray Evaluation of Antimicrobial Resistance and Virulence of Escherichia coli Isolates from Portuguese Poultry. Antibiotics 2016, 5, 4. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics5010004
Mendonça N, Figueiredo R, Mendes C, Card RM, Anjum MF, Da Silva GJ. Microarray Evaluation of Antimicrobial Resistance and Virulence of Escherichia coli Isolates from Portuguese Poultry. Antibiotics. 2016; 5(1):4. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics5010004
Chicago/Turabian StyleMendonça, Nuno, Rui Figueiredo, Catarina Mendes, Roderick M. Card, Muna F. Anjum, and Gabriela Jorge Da Silva. 2016. "Microarray Evaluation of Antimicrobial Resistance and Virulence of Escherichia coli Isolates from Portuguese Poultry" Antibiotics 5, no. 1: 4. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics5010004





