The Perfect Bacteriophage for Therapeutic Applications—A Quick Guide
Abstract
:1. Phage Therapy: A Major Comeback with Minor Setbacks
2. In Search of New Phages
3. Desirable Traits in Phages with Antimicrobial Potential
3.1. Specific but Not Too Specific
3.2. Virulent Phages Only, Please
3.3. Safe for Human Health and the Environment
3.4. Stability
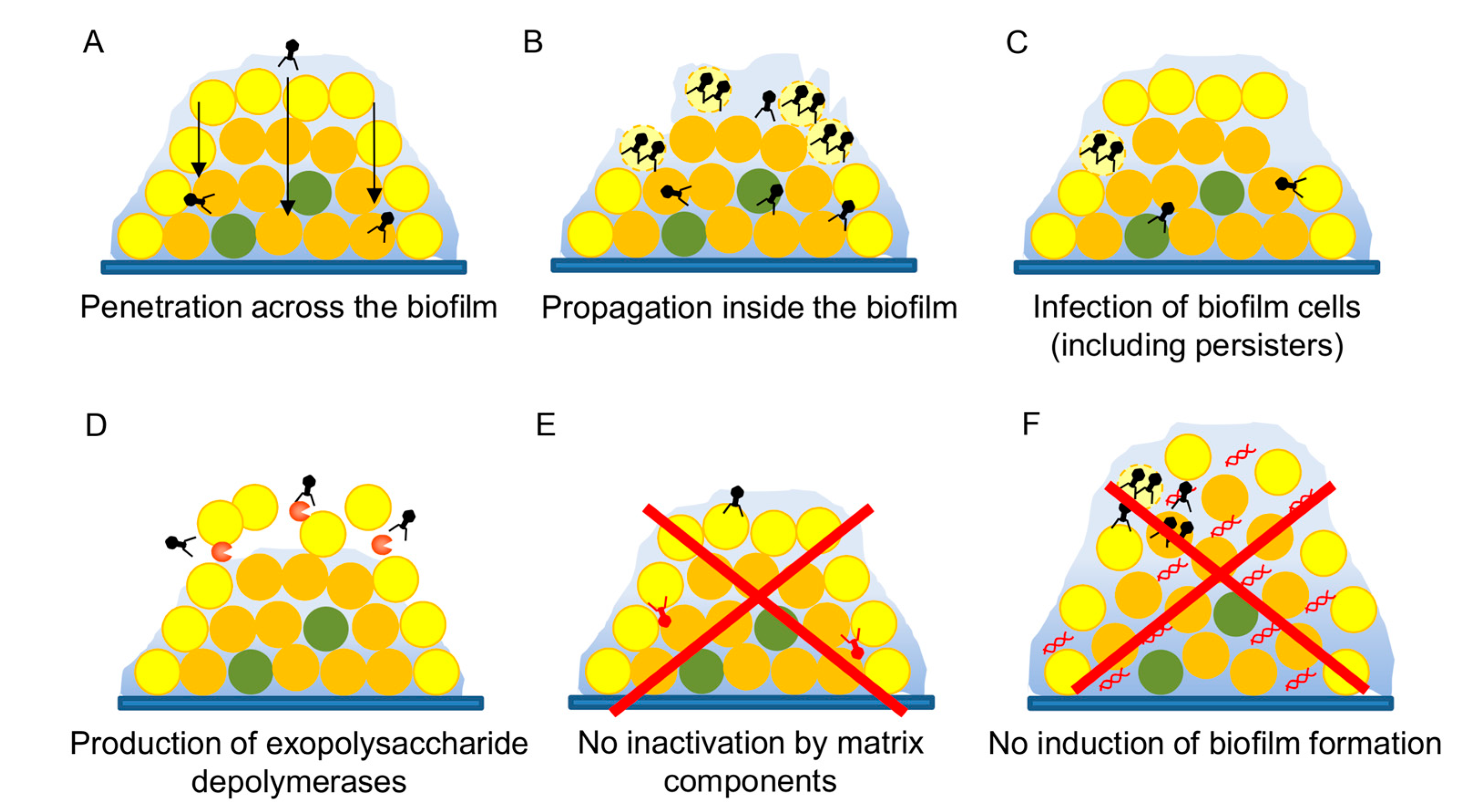
3.5. Antibiofilm Potential
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- D’Hérelle, F. Sur un microbe invisible antagoniste des bacilles dysentériques. C. R. Acad. Sci. 1917, 165, 373–375. [Google Scholar]
- Twort, F. An investigation on the nature of ultra-microscopic viruses. Lancet 1915, 186, 1241–1243. [Google Scholar] [CrossRef]
- D’Hérelle, F.; Smith, G.H. The Bacteriophage and Its Clinical Application; Charles, C., Ed.; Thomas: Springfield, IL, USA, 1930. [Google Scholar]
- Debarbieux, L.; Pirnay, J.P.; Verbeken, G.; de Vos, D.; Merabishvili, M.; Huys, I.; Patey, O.; Schoonjans, D.; Vaneechoutte, M.; Zizi, M.; et al. A bacteriophage journey at the European Medicines Agency. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- World Health Organization. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Gordillo-Altamirano, F.L.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Furfaro, L.L.; Payne, M.S.; Chang, B.J. Bacteriophage therapy: Clinical trials and regulatory hurdles. Front. Cell Infect. Microbiol. 2018, 8, 376. [Google Scholar] [CrossRef]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage applications for food production and processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef]
- Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Application of bacteriophages in the agro-food sector: A long way toward approval. Front. Cell Infect. Microbiol. 2018, 8, 296. [Google Scholar] [CrossRef]
- Endersen, L.; O’Mahony, J.; Hill, C.; Ross, R.P.; McAuliffe, O.; Coffey, A. Phage therapy in the food industry. Annu. Rev. Food Sci. Technol. 2014, 5, 327–349. [Google Scholar] [CrossRef]
- McCallin, S.; Sacher, J.C.; Zheng, J.; Chan, B.K. Current state of compassionate phage therapy. Viruses 2019, 11, 343. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Vos, M.; Birkett, P.J.; Birch, E.; Griffiths, R.I.; Buckling, A. Local adaptation of bacteriophages to their bacterial hosts in soil. Science 2009, 325, 833. [Google Scholar] [CrossRef] [PubMed]
- D’Herelle, F. Le Phénomène de la Guérison des Maladies Infectieuses; Masson: Paris, France, 1938. [Google Scholar]
- O’Flynn, G.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.L.; Petrovski, S.; Dyson, Z.A.; Seviour, R.; Tucci, J. The formulation of bacteriophage in a semi solid preparation for control of Propionibacterium acnes growth. PLoS ONE 2016, 11, e0151184. [Google Scholar] [CrossRef] [PubMed]
- Tylenda, C.A.; Calvert, C.; Kolenbrander, P.E.; Tylenda, A. Isolation of Actinomyces bacteriophage from human dental plaque. Infect. Immun. 1985, 49, 1–6. [Google Scholar] [PubMed]
- Latz, S.; Wahida, A.; Arif, A.; Häfner, H.; Hoß, M.; Ritter, K.; Horz, H.P. Preliminary survey of local bacteriophages with lytic activity against multi-drug resistant bacteria. J. Basic Microbiol. 2016, 56, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Weber-Dąbrowska, B.; JońCzyk-Matysiak, E.; Żaczek, M.; Łobocka, M.; Łusiak-Szelachowska, M.; Górski, A. Bacteriophage procurement for therapeutic purposes. Front. Microbiol. 2016, 7, 1177. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Vervaet, C.; Pirnay, J.-P.; de Vos, D.; Verbeken, G.; Mast, J.; Chanishvili, N.; Vaneechoutte, M. Stability of Staphylococcus aureus phage ISP after freeze-drying (lyophilization). PLoS ONE 2013, 8, e68797. [Google Scholar] [CrossRef]
- Ul Haq, I.; Chaudhry, W.N.; Andleeb, S.; Qadr, I.I. Isolation and partial characterization of a virulent bacteriophage IHQ1 specific for Aeromonas punctata from stream water. Microb. Ecol. 2011, 63, 954–963. [Google Scholar] [CrossRef]
- Weber-Dąbrowska, B.; Żaczek, M.; Dziedzic, B.; Łusiak-Szelachowska, M.; Kiejzik, M.; Górski, A.; Gworek, B.; Wierzbicki, K.; Eymontt, A. Bacteriophages in green biotechnology—The utilization of drinking water. In Industrial, Medical and Environmental Applications of Microorganisms: Current Status and Trends; Wageningen Academic Publishers: Wageningen, The Netherlands, 2014; pp. 500–504. [Google Scholar]
- Vongkamjan, K.; Switt, A.M.; den Bakker, H.C.; Fortes, E.D.; Wiedmann, M. Silage collected from dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Appl. Environ. Microbiol. 2012, 78, 8666–8675. [Google Scholar] [CrossRef] [PubMed]
- Loś, J.M.; Golec, P.; Wegrzyn, G.; Wegrzyn, A.; Loś, M. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl. Environ. Microbiol. 2008, 74, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Harjai, K.; Chhibber, S. Methicillin-resistant Staphylococcus aureus phage plaque size enhancement using sublethal concentrations of antibiotics. Appl. Environ. Microbiol. 2012, 78, 8227–8233. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, M.R.; Balaa, M.F. Enhanced contrast of bacteriophage plaques in Salmonella with ferric ammonium citrate and sodium thiosulfate (FACST) and tetrazolium red (TZR). J. Microbiol. Methods 2006, 65, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Casey, E.; van Sinderen, D.; Mahony, J. In vitro characteristics of phages to guide ‘real life’ phage therapy suitability. Viruses 2018, 10, 163. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Liu, X.; Li, Y.; Han, W.; Lei, L.; Yang, Y.; Zhao, H.; Gao, Y.; Song, J.; Lu, R.; et al. A method for generation phage cocktail with great therapeutic potential. PLoS ONE 2012, 7, e31698. [Google Scholar] [CrossRef]
- Kawecki, T.J.; Ebert, D. Conceptual issues of local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 39. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Vandenheuvel, D.; Martínez, B.; Rodríguez, A.; Lavigne, R.; García, P. Two phages, phiIPLA-RODI and phiIPLA-C1C, lyse mono- and dual-species staphylococcal biofilms. Appl. Environ. Microbiol. 2015, 81, 3336–3348. [Google Scholar] [CrossRef]
- El Haddad, L.; Ben Abdallah, N.; Plante, P.L.; Dumaresq, J.; Katsarava, R.; Labrie, S.; Corbeil, J.; St-Gelais, D.; Moineau, S. Improving the safety of Staphylococcus aureus polyvalent phages by their production on a Staphylococcus xylosus strain. PLoS ONE 2014, 9, e102600. [Google Scholar] [CrossRef]
- Hamdi, S.; Rousseau, G.M.; Labrie, S.J.; Tremblay, D.M.; Kourda, R.S.; Ben Slama, K.; Moineau, S. Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci. Rep. 2017, 7, 40349. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Meaden, S. Understanding bacteriophage specificity in natural microbial communities. Viruses 2013, 5, 806. [Google Scholar] [CrossRef] [PubMed]
- Poullain, V.; Gandon, S.; Brockhurst, M.A.; Buckling, A.; Hochberg, M.E. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution 2008, 62, 1–11. [Google Scholar] [CrossRef]
- Duffy, S.; Turner, P.E.; Burch, C.L. Pleiotropic costs of niche expansion in the RNA bacteriophage Φ6. Genetics 2006, 172, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Avrani, S.; Schwartz, D.A.; Lindell, D. Virus-host swinging party in the oceans: Incorporating biological complexity into paradigms of antagonistic coexistence. Mob. Gent. Elem. 2012, 2, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Phage Receptor Database. Available online: https://phred.herokuapp.com (accessed on 18 July 2019).
- Buckling, A.; Brockhurst, M. Bacteria-virus coevolution. Adv. Exp. Med. Biol. 2012, 751, 347–370. [Google Scholar] [CrossRef]
- Tsai, R.; Corrêa, I.R.; Xu, M.Y.; Xu, S.Y. Restriction and modification of deoxyarchaeosine (dG+)-containing phage 9 g DNA. Sci. Rep. 2017, 7, 8348. [Google Scholar] [CrossRef]
- Šimoliunas, E.; Šimoliuniene, M.; Kaliniene, L.; Zajanckauskaite, A.; Skapas, M.; Mešky, S.R.; Kaupinis, A.; Valius, M.; Truncaite, L. Pantoea bacteriophage vB_PagS_Vid5: A low-temperature siphovirus that harbors a cluster of genes involved in the biosynthesis of archaeosine. Viruses 2018, 10, 583. [Google Scholar] [CrossRef]
- Ngazoa-Kakou, S.; Shao, Y.; Rousseau, G.M.; Addablah, A.A.; Tremblay, D.M.; Hutinet, G.; Lemire, N.; Plante, P.L.; Corbeil, J.; Koudou, A.; et al. Complete genome sequence of Escherichia coli siphophage BRET. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef]
- Hobbs, Z.; Abedon, S.T. Diversity of phage infection types and associated terminology: The problem with ‘lytic or lysogenic’. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Fernández, L.; Rodríguez, A.; García, P. Practical method for isolation of phage deletion mutants. Methods Protoc. 2018, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef] [PubMed]
- Keen, E.C.; Bliskovsky, V.V.; Malagon, F.; Baker, J.D.; Prince, J.S.; Klaus, J.S.; Adhya, S.L. Novel “superspreader” bacteriophages promote horizontal gene transfer by transformation. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Baig, A.; Colom, J.; Barrow, P.; Schouler, C.; Moodley, A.; Lavigne, R.; Atterbury, R. Biology and genomics of an historic therapeutic Escherichia coli bacteriophage collection. Front. Microbiol. 2017, 8, 1652. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.J.; Hyman, P. Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 2010, 11, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Speck, P.; Smithyman, A. Safety and efficacy of phage therapy via the intravenous route. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Salmon, G.G., Jr.; Symonds, M. Staphage lysate therapy in chronic staphylococcal infections. J. Med. Soc. N. J. 1963, 60, 188–193. [Google Scholar] [PubMed]
- Meaden, S.; Koskella, B. Exploring the risks of phage application in the environment. Front. Microbiol. 2013, 4, 358. [Google Scholar] [CrossRef] [Green Version]
- Regulski, K.; Champion-Arnaud, P.; Gabard, J. Bacteriophage manufacturing: From early twentieth-century processes to current GMP. In Bacteriophages; Harper, D., Ed.; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Jurač, K.; Nabergoj, D.; Podgornik, A. Bacteriophage production processes. Appl. Microbiol. Biotechnol. 2019, 103, 685–694. [Google Scholar] [CrossRef]
- Pirnay, J.P.; Merabishvili, M.; Van Raemdonck, H.; De Vos, D.; Verbeken, G. Bacteriophage production in compliance with regulatory requirements. Methods Mol. Biol. 2018, 1693, 233–252. [Google Scholar] [CrossRef] [PubMed]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages—Review. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W.; Tremblay, D.; Moineau, S. Long-term bacteriophage preservation. WFCC Newslett. 2004, 38, 35–40. [Google Scholar]
- González-Menéndez, E.; Fernández, L.; Gutiérrez, D.; Rodríguez, A.; Martínez, B.; García, P. Comparative analysis of different preservation techniques for the storage of Staphylococcus phages aimed for the industrial development of phage-based antimicrobial products. PLoS ONE 2018, 13, e0205728. [Google Scholar] [CrossRef] [PubMed]
- Hubalek, Z. Protectants used in the cryopreservation of microorganisms. Cryobiology 2003, 46, 205–229. [Google Scholar] [CrossRef]
- Golec, P.; Dabrowski, K.; Hejnowicz, M.S.; Gozdek, A.; Loś, J.M.; Wegrzyn, G.; Lobocka, M.B.; Loś, M. A reliable method for storage of tailed phages. J. Microbiol. Methods 2011, 84, 486–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Menéndez, E.; Fernández, L.; Gutiérrez, D.; Pando, D.; Martínez, B.; Rodríguez, A.; García, P. Strategies to encapsulate the Staphylococcus aureus bacteriophage phiIPLA-RODI. Viruses 2018, 10, 495. [Google Scholar] [CrossRef] [PubMed]
- Bodier-Montagutelli, E.; Morello, E.; L’Hostis, G.; Guillon, A.; Dalloneau, E.; Respaud, R.; Pallaoro, N.; Blois, H.; Vecellio, L.; Gabard, J.; et al. Inhaled phage therapy: A promising and challenging approach to treat bacterial respiratory infections. Expert Opin. Drug Deliv. 2017, 14, 959–972. [Google Scholar] [CrossRef]
- Leung, S.S.Y.; Parumasivam, T.; Gao, F.G.; Carter, E.A.; Carrigy, N.B.; Vehring, R.; Finlay, W.H.; Morales, S.; Britton, W.J.; Kutter, E.; et al. Effects of storage conditions on the stability of spray dried, inhalable bacteriophage powders. Int. J. Pharm. 2017, 521, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Leung, S.S.; Parumasivam, T.; Gao, F.G.; Carrigy, N.B.; Vehring, R.; Finlay, W.H.; Morales, S.; Britton, W.J.; Kutter, E.; Chan, H.K. Production of inhalation phage powders using spray freeze drying and spray drying techniques for treatment of respiratory infections. Pharm. Res. 2016, 33, 1486–1496. [Google Scholar] [CrossRef]
- Ma, Y.; Pacan, J.C.; Wang, Q.; Xu, Y.; Huang, X.; Korenevsky, A.; Sabour, P.M. Microencapsulation of bacteriophage Felix O1 into chitosan-alginate microspheres for oral delivery. Appl. Environ. Microbiol. 2008, 74, 4799–4805. [Google Scholar] [CrossRef] [PubMed]
- Singla, S.; Harjai, K.; Katare, O.P.; Chhibber, S. Bacteriophage-loaded nanostructured lipid carrier: Improved pharmacokinetics mediates effective resolution of Klebsiella pneumoniae—Induced lobar pneumonia. J. Infect. Dis. 2015, 212, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Nieth, A.; Verseux, C.; Barnert, S.; Süss, R.; Römer, W. A first step toward liposome-mediated intracellular bacteriophage therapy. Expert Opin. Drug Deliv. 2015, 12, 1411–1424. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.L.; Costa, A.R.; Santos, J.F.; Siliakus, M.F.; van Lent, J.W.; Kengen, S.W.; Azeredo, J.; Kluskens, L.D. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci. Rep. 2016, 6, 39235. [Google Scholar] [CrossRef] [Green Version]
- Esteban, P.P.; Alves, D.R.; Enright, M.C.; Bean, J.E.; Gaudion, A.; Jenkins, A.T.; Young, A.E.; Arnot, T.C. Enhancement of the antimicrobial properties of bacteriophage-K via stabilization using oil-in-water nano-emulsions. Biotechnol. Prog. 2014, 30, 932–944. [Google Scholar] [CrossRef] [Green Version]
- Esteban, P.P.; Jenkins, A.T.; Arnot, T.C. Elucidation of the mechanisms of action of bacteriophage K/nano-emulsion formulations against Staphylococcus aureus via measurement of particle size and zeta potential. Colloids Surf. B Biointerfaces 2016, 139, 87–94. [Google Scholar] [CrossRef]
- Merabishvili, M.; Monserez, R.; van Belleghem, J.; Rose, T.; Jennes, S.; de Vos, D.; Verbeken, G.; Vaneechoutte, M.; Pirnay, J.P. Stability of bacteriophages in burn wound care products. PLoS ONE 2017, 12, e0182121. [Google Scholar] [CrossRef]
- Majewska, J.; Beta, W.; Lecion, D.; Hodyra-Stefaniak, K.; Kłopot, A.; Kaźmierczak, Z.; Miernikiewicz, P.; Piotrowicz, A.; Ciekot, J.; Owczarek, B.; et al. Oral application of T4 phage induces weak antibody production in the gut and in the blood. Viruses 2015, 7, 2845. [Google Scholar] [CrossRef]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [Green Version]
- Kucharewicz-Krukowska, A.; Slopek, S. Immunogenic effect of bacteriophage in patients subjected to phage therapy. Arch. Immunol. Ther. Exp. 1987, 35, 553–561. [Google Scholar]
- Kim, K.P.; Cha, J.D.; Jang, E.H.; Klumpp, J.; Hagens, S.; Hardt, W.D.; Lee, K.Y.; Loessner, M.J. PEGylation of bacteriophages increases blood circulation time and reduces T-helper type 1 immune response. Microb. Biotechnol. 2008, 1, 247–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Núñez, C.; Reffuveille, F.; Fernández, L.; Hancock, R.E.W. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Bacteriophages and their enzymes in biofilm control. Curr. Pharm. Des. 2015, 21, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Geredew Kifelew, L.; Mitchell, J.G.; Speck, P. Mini-review: Efficacy of lytic bacteriophages on multispecies biofilms. Biofouling 2019, 35, 472–481. [Google Scholar] [CrossRef] [PubMed]
- González, S.; Fernández, L.; Campelo, A.B.; Gutiérrez, D.; Martínez, B.; Rodríguez, A.; García, P. The behavior of Staphylococcus aureus dual-species biofilms treated with bacteriophage phiIPLA-RODI depends on the accompanying microorganism. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Brosh, Y.; Gelman, D.; Coppenhagen-Glazer, S.; Beyth, S.; Poradosu-Cohen, R.; Que, Y.A.; Beyth, N.; Hazan, R. Targeting Enterococcus faecalis biofilms with phage therapy. Appl. Environ. Microbiol. 2015, 81, 2696–2705. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Qu, K.; Tan, D.; Li, X.; Wang, L.; Cong, C.; Xiu, Z.; Xu, Y. Isolation and characterization of a bacteriophage and its potential to disrupt multi-drug resistant Pseudomonas aeruginosa biofilms. Microb. Pathog. 2019, 128, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Naknaen, A.; Pomwised, R.; Voravuthikunchai, S.P.; Smith, D.R. Isolation and characterization of Siphoviridae phage infecting extensively drug-resistant Acinetobacter baumannii and evaluation of therapeutic efficacy In Vitro and In Vivo. J. Med. Microbiol. 2019, 68, 1096–1108. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, Y.; Xu, J.; Yu, X.; Huang, X.; Liu, G.; Liu, X. Identification of novel bacteriophage vB_EcoP-EG1 with lytic activity against planktonic and biofilm forms of uropathogenic Escherichia coli. Appl. Microbiol. Biotechnol. 2019, 103, 315–326. [Google Scholar] [CrossRef]
- Briandet, R.; Lacroix-Gueu, P.; Renault, M.; Lecart, S.; Meylheuc, T.; Bidnenko, E.; Steenkeste, K.; Bellon-Fontaine, M.N.; Fontaine-Aupart, M.P. Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Appl. Environ. Microbiol. 2008, 74, 2135–2143. [Google Scholar] [CrossRef] [PubMed]
- González, S.; Fernández, L.; Gutiérrez, D.; Campelo, A.B.; Rodríguez, A.; García, P. Analysis of different parameters affecting diffusion, propagation and survival of staphylophages in bacterial biofilms. Front. Microbiol. 2018, 9, 2348. [Google Scholar] [CrossRef] [PubMed]
- Tkhilaishvili, T.; Lombardi, L.; Klatt, A.B.; Trampuz, A.; Di Luca, M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 52, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.K.; Collins, J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA 2007, 104, 11197–11202. [Google Scholar] [CrossRef] [Green Version]
- Pei, R.; Lamas-Samanamud, G.R. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl. Environ. Microbiol. 2014, 80, 5340–5348. [Google Scholar] [CrossRef]
- Fernández, L.; Rodríguez, A.; García, P. Phage or foe: An insight into the impact of viral predation on microbial communities. ISME J. 2018, 12, 1171–1179. [Google Scholar] [CrossRef]
- Fernández, L.; González, S.; Campelo, A.B.; Martínez, B.; Rodríguez, A.; García, P. Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus. Sci. Rep. 2017, 7, 40965. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Tufenkji, N.; van de Ven, T.G. Formation of biofilms under phage predation: Considerations concerning a biofilm increase. Biofouling 2013, 29, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Dahl, A.; Middelboe, M. Vibriophages differentially influence biofilm formation by Vibrio anguillarum strains. Appl. Environ. Microbiol. 2015, 81, 4489–4497. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández, L.; Gutiérrez, D.; García, P.; Rodríguez, A. The Perfect Bacteriophage for Therapeutic Applications—A Quick Guide. Antibiotics 2019, 8, 126. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics8030126
Fernández L, Gutiérrez D, García P, Rodríguez A. The Perfect Bacteriophage for Therapeutic Applications—A Quick Guide. Antibiotics. 2019; 8(3):126. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics8030126
Chicago/Turabian StyleFernández, Lucía, Diana Gutiérrez, Pilar García, and Ana Rodríguez. 2019. "The Perfect Bacteriophage for Therapeutic Applications—A Quick Guide" Antibiotics 8, no. 3: 126. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics8030126





