The Application of Ribosome Engineering to Natural Product Discovery and Yield Improvement in Streptomyces
Abstract
:1. Introduction
2. Application of Ribosome Engineering to Increase Antibiotic Production
2.1. Single Drug Resistance Mutation
2.2. Combinations of Drug Resistance Mutation
2.3. Combination of Traditional Mutagenesis and Ribosome Engineering
2.4. Combination of Genome Shuffling and Ribosome Engineering
2.5. Overexpression of Ribosome Recycling Factor
2.6. The Application of Ribosome Engineering in Other Bacteria and Fungi
3. Discovery of New Natural Products Using Ribosome Engineering
4. Possible Mechanism of Action of Ribosome Engineering
4.1. The Stringent Response, ppGpp, and Ribosome Engineering
4.2. Ribosome Stability, Recycling, and Streptomycin-Resistance
4.3. Sublethal Concentrations of Different Antibiotics and Ribosome Engineering
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Demain, A.L.; Sánchez, S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, B. A new golden age of natural products drug discovery. Cell 2015, 163, 1297–1300. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Deng, Z.; Liu, T. Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab. Eng. 2018, 50, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Charusanti, P.; Musiol-Kroll, E.M.; Jiang, X.; Tong, Y.; Kim, H.U.; Lee, S.Y. Metabolic engineering of antibiotic factories: New tools for antibiotic production in actinomycetes. Trends Biotechnol. 2015, 33, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Heitman, J.; Movva, N.; Hall, M. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.; Colombi, M.; Moroni, C.; Hall, M.N. Rapamycin passes the torch: A new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 2011, 10, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Hong, S.Y.; Wang, J.; Rehan, S.; Liu, W.; Peng, H.; Das, M.; Li, W.; Bhat, S.; Peiffer, B.; et al. Rapamycin-inspired macrocycles with new target specificity. Nat. Chem. 2019, 11, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Ishikawa, J.; Hanamoto, A.; Shinose, M.; Kikuchi, H.; Shiba, T.; Sakaki, Y.; Hattori, M.; Ōmura, S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003, 21, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Yan, X.; Chen, J.-J.; Adhikari, A.; Yang, D.; Crnovcic, I.; Wang, N.; Chang, C.-Y.; Rader, C.; Shen, B. Genome mining of Micromonospora yangpuensis DSM 45577 as a producer of an anthraquinone-fused enediyne. Org. Lett. 2017, 19, 6192–6195. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, J.D.; Yan, X.; Shen, B. Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery. J. Ind. Microbiol. Biotechnol. 2016, 43, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Xu, Z.; Guo, Z.; Hindra; Ma, M.; Yang, D.; Zhou, H.; Gansemans, Y.; Zhu, X.; Huang, Y.; et al. Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc. Natl. Acad. Sci. USA 2017, 114, E11131–E11140. [Google Scholar] [CrossRef] [PubMed]
- Essential Medicines. Available online: http://www.who.int/topics/essential_medicines (accessed on 28 July 2019).
- Shima, J.; Hesketh, A.; Okamoto, S.; Kawamoto, S.; Ochi, K. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 1996, 178, 7276–7284. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K. Insights into microbial cryptic gene activation and strain improvement: Principle, application and technical aspects. J. Antibiot. 2017, 70, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Genet. 2016, 14, 135–149. [Google Scholar] [CrossRef]
- Nielsen, J.; Keasling, J.D. Engineering cellular metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef]
- Bose, J.L. Chemical and UV mutagenesis. Methods Mol. Biol. 2016, 1373, 111–115. [Google Scholar]
- Gong, J.; Zheng, H.; Wu, Z.; Chen, T.; Zhao, X. Genome shuffling: Progress and applications for phenotype improvement. Biotechnol. Adv. 2009, 27, 996–1005. [Google Scholar] [CrossRef]
- Magocha, T.A.; Zabed, H.; Yang, M.; Yun, J.; Zhang, H.; Qi, X. Improvement of industrially important microbial strains by genome shuffling: Current status and future prospects. Bioresour. Technol. 2018, 257, 281–289. [Google Scholar] [CrossRef]
- Xu, M.; Wright, G.D. Heterologous expression-facilitated natural products’ discovery in actinomycetes. J. Ind. Microbiol. Biotechnol. 2018, 46, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Jeschek, M.; Gerngross, D.; Panke, S. Combinatorial pathway optimization for streamlined metabolic engineering. Curr. Opin. Biotechnol. 2017, 47, 142–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, D.G.; Glass, J.I.; Lartigue, C.; Noskov, V.N.; Chuang, R.-Y.; Algire, M.A.; Benders, G.A.; Montague, M.G.; Ma, L.; Moodie, M.M.; et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, A.; Ochi, K. A novel method for improving Streptomyces coelicolor A3(2) for production of actinorhodin by introduction of rpsL (encoding ribosomal protein S12) mutations conferring resistance to streptomycin. J. Antibiot. 1997, 50, 532–535. [Google Scholar] [CrossRef] [PubMed]
- Okamoto-Hosoya, Y.; Sato, T.-A.; Ochi, K. Resistance to paromomycin is conferred by rpsL mutations, accompanied by an enhanced antibiotic production in Streptomyces coelicolor A3(2). J. Antibiot. 2000, 53, 1424–1427. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Ochi, K. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Environ. Microbiol. 2001, 67, 1885–1892. [Google Scholar] [CrossRef]
- Xu, J.; Tozawa, Y.; Lai, C.; Hayashi, H.; Ochi, K. A rifampicin resistance mutation in the rpoB gene confers ppGpp-independent antibiotic production in Streptomyces coelicolor A3(2). Mol. Genet. Genom. 2002, 268, 179–189. [Google Scholar] [CrossRef]
- Wang, G.; Hosaka, T.; Ochi, K. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl. Environ. Microbiol. 2008, 74, 2834–2840. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Komatsu, M.; Okamoto, S.; Tokuyama, S.; Kaji, A.; Ikeda, H.; Ochi, K. Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl. Environ. Microbiol. 2009, 75, 4919–4922. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Fujiwara, T.; Ochi, K.; Hosaka, T. Development of the ability to produce secondary metabolites in Streptomyces through the acquisition of erythromycin resistance. J. Antibiot. 2012, 65, 323–326. [Google Scholar] [CrossRef]
- Tanaka, Y.; Kasahara, K.; Hirose, Y.; Murakami, K.; Kugimiya, R.; Ochi, K. Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in actinomycetes. J. Bacteriol. 2013, 195, 2959–2970. [Google Scholar] [CrossRef]
- Hosoya, Y.; Okamoto, S.; Muramatsu, H.; Ochi, K. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob. Agents Chemother. 1998, 42, 2041–2047. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.-F.; Zhang, Q.; Zhu, B.-Q. Enhanced antibiotic production by inducing low level of resistance to gentamicin. Chin. J. Nat. Med. 2008, 6, 146–152. [Google Scholar] [CrossRef]
- Li, L.; Guo, J.; Wen, Y.; Chen, Z.; Song, Y.; Li, J. Overexpression of ribosome recycling factor causes increased production of avermectin in Streptomyces avermitilis strains. J. Ind. Microbiol. Biotechnol. 2010, 37, 673–679. [Google Scholar] [CrossRef]
- Lv, X.A.; Jin, Y.Y.; Li, Y.D.; Zhang, H.; Liang, X.L. Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin. Appl. Microbiol. Biotechnol. 2013, 97, 641–648. [Google Scholar] [CrossRef]
- Gomez-Escribano, J.P.; Bibb, M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, T.; Liu, Q.; Huang, Y.; Hu, C.; Liao, G. Improvement of daptomycin production in Streptomyces roseosporus through the acquisition of pleuromutilin resistance. BioMed. Res. Int. 2013, 2013, 1–6. [Google Scholar]
- Yu, G.; Hui, M.; Li, R.; Chen, L.; Tian, H.; Wang, L. Enhancement of daptomycin production by the method of combining ribosome engineering and genome shuffling in Streptomyces roseosporus. Appl. Biochem. Microbiol. 2018, 54, 611–615. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Y.; Liu, Q.; Huang, Y.; Hu, C.; Liao, G. Improvement of A21978C production in Streptomyces roseosporus by reporter-guided rpsL mutation selection. J. Appl. Microbiol. 2012, 112, 1095–1101. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, H.; Xu, S.; Wang, B.; Ju, J.; Tan, H.; Li, W. Activation and enhancement of fredericamycin A production in deep sea-derived Streptomyces somaliensis SCSIO ZH66 by using ribosome engineering and response surface methodology. Microb. Cell Factories 2015, 14, 1039. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-J.; Wang, X.-C.; Xiang, W.-S. Improvement of milbemycin-producing Streptomyces bingchenggensis by rational screening of ultraviolet- and chemically induced mutants. World J. Microbiol. Biotechnol. 2009, 25, 1051–1056. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, D.; Li, Y.; Zhang, F.; Wang, C.; Liang, X. Genome shuffling and ribosome engineering of Streptomyces actuosus for high-yield nosiheptide production. Appl. Biochem. Biotechnol. 2014, 173, 1553–1563. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, Z.; Ma, Z.; Bechthold, A.; Yu, X. Sequential improvement of rimocidin production in Streptomyces rimosus M527 by introduction of cumulative drug-resistance mutations. J. Ind. Microbiol. Biotechnol. 2019, 46, 697–708. [Google Scholar] [CrossRef]
- Tamehiro, N.; Hosaka, T.; Xu, J.; Hu, H.; Otake, N.; Ochi, K. Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl. Environ. Microbiol. 2003, 69, 6412–6417. [Google Scholar] [CrossRef]
- Fukuda, K.; Tamura, T.; Ito, H.; Yamamoto, S.; Ochi, K.; Inagaki, K. Production improvement of antifungal, antitrypanosomal nucleoside sinefungin by rpoB mutation and optimization of resting cell system of Streptomyces incarnatus NRRL 8089. J. Biosci. Bioeng. 2010, 109, 459–465. [Google Scholar] [CrossRef]
- Yan, X.; Ge, H.; Huang, T.; Hindra; Yang, D.; Teng, Q.; Crnovčić, I.; Li, X.; Rudolf, J.D.; Lohman, J.R.; et al. Strain prioritization and genome mining for enediyne natural products. mBio 2016, 7, 02104–02116. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Pan, J.; Wang, Z.; Yan, X.; Yang, D.; Zhu, X.; Shen, B.; Duan, Y.; Huang, Y. Ribosome engineering and fermentation optimization leads to overproduction of tiancimycin A, a new enediyne natural product from Streptomyces sp. CB03234. J. Ind. Microbiol. Biotechnol. 2018, 45, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Jiang, C.; Zhang, F.; Huang, R.; Yi, L.; Huang, Y.; Yan, X.; Duan, Y.; Zhu, X. Streptomycin-induced ribosome engineering complemented with fermentation optimization for enhanced production of 10-membered enediynes tiancimycin-A and tiancimycin-D. Biotechnol. Bioeng. 2019, 116, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Tao, L.; Bechthold, A.; Shentu, X.; Bian, Y.; Yu, X. Overexpression of ribosome recycling factor is responsible for improvement of nucleotide antibiotic-toyocamycin in Streptomyces diastatochromogenes 1628. Appl. Microbiol. Biotechnol. 2014, 98, 5051–5058. [Google Scholar] [CrossRef]
- Ma, Z.; Luo, S.; Xu, X.; Bechthold, A.; Yu, X. Characterization of representative rpoB gene mutations leading to a significant change in toyocamycin production of Streptomyces diastatochromogenes 1628. J. Ind. Microbiol. Biotechnol. 2016, 43, 463–471. [Google Scholar] [CrossRef]
- Tong, Q.-Q.; Zhou, Y.-H.; Chen, X.-S.; Wu, J.-Y.; Wei, P.; Yuan, L.-X.; Yao, J.-M. Genome shuffling and ribosome engineering of Streptomyces virginiae for improved virginiamycin production. Bioprocess Biosyst. Eng. 2018, 41, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Kong, J.; Yang, H.; Huang, R.; Huang, Y.; Yang, D.; Shen, B.; Duan, Y. Strain improvement by combined UV mutagenesis and ribosome engineering and subsequent fermentation optimization for enhanced 6’-deoxy-bleomycin Z production. Appl. Microbiol. Biotechnol. 2018, 102, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Tojo, S.; Tanaka, Y.; Ochi, K. Activation of antibiotic production in Bacillus spp. by cumulative drug resistance mutations. Antimicrob. Agents Chemother. 2015, 59, 7799–7804. [Google Scholar] [CrossRef] [PubMed]
- Carata, E.; Peano, C.; Tredici, S.M.; Ferrari, F.; Tala, A.; Corti, G.; Bicciato, S.; De Bellis, G.; Alifano, P. Phenotypes and gene expression profiles of Saccharopolyspora erythraea rifampicin-resistant (rif) mutants affected in erythromycin production. Microb Cell Fact 2009, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Beltrametti, F.; Rossi, R.; Selva, E.; Marinelli, F. Antibiotic production improvement in the rare actinomycete Planobispora rosea by selection of mutants resistant to the aminoglycosides streptomycin and gentamycin and to rifamycin. J. Ind. Microbiol. Biotechnol. 2006, 33, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Zhu, Y.; Liu, S.; Cai, C.; Xu, P. Screening of high yield norvancomycin producing strain by streptomycin and rifampicin resistant mutation. Chin. J. Antibiot. 2006, 31, 243–246. [Google Scholar]
- Ahmetagic, A.; Pemberton, J.M. Antibiotic resistant mutants of Escherichia coli K12 show increases in heterologous gene expression. Plasmid 2011, 65, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Funane, K.; Tanaka, Y.; Hosaka, T.; Murakami, K.; Miyazaki, T.; Shiwa, Y.; Gibu, S.; Inaoka, T.; Kasahara, K.; Fujita, N.; et al. Combined drug resistance mutations substantially enhance enzyme production in Paenibacillus agaridevorans. J. Bacteriol. 2018, 200, JB.00188-18. [Google Scholar] [CrossRef]
- Kurosawa, K.; Hosaka, T.; Tamehiro, N.; Inaoka, T.; Ochi, K. Improvement of alpha-amylase production by modulation of ribosomal component protein S12 in Bacillus subtilis 168. Appl. Environ. Microbiol. 2006, 72, 71–77. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, X.; Bai, F. Production of xylanase by an alkaline-tolerant marine-derived Streptomyces viridochromogenes strain and improvement by ribosome engineering. Appl. Microbiol. Biotechnol. 2013, 97, 4361–4368. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kasahara, K.; Izawa, M.; Ochi, K. Applicability of ribosome engineering to vitamin B12 production by Propionibacterium shermanii. Biosci. Biotechnol. Biochem. 2017, 81, 1636–1641. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Wang, L.; Gao, Y.; Xiang, J.; Chen, X.; Mao, Z. Breeding and fermentation performance of a high-yield ε-poly-L-lysine producing strain. Acta Microbiol. Sin. 2016, 56, 1450–1458. [Google Scholar]
- Liu, Y.-J.; Chen, X.-S.; Zhao, J.-J.; Pan, L.; Mao, Z.-G. Development of microtiter plate culture method for rapid screening of ε-Poly-L-lysine-producing strains. Appl. Biochem. Biotechnol. 2017, 183, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, X.; Wu, G.; Li, S.; Zeng, X.; Ren, X.; Tang, L.; Mao, Z. Enhanced ε-poly-L-lysine production by inducing double antibiotic-resistant mutations in Streptomyces albulus. Bioprocess Biosyst. Eng. 2017, 40, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Zhao, J.; Li, Q.; Mao, Z. Improvement of ε-poly-L-lysine production of Streptomyces albulus by continuous introduction of streptomycin resistance. Process. Biochem. 2019, 82, 10–18. [Google Scholar] [CrossRef]
- Zhao, J.; Hu, X.; Li, Y.; Wang, X. Overexpression of ribosome elongation factor G and recycling factor increases L-isoleucine production in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2015, 99, 4795–4805. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kasahara, K.; Hirose, Y.; Morimoto, Y.; Izawa, M.; Ochi, K. Enhancement of butanol production by sequential introduction of mutations conferring butanol tolerance and streptomycin resistance. J. Biosci. Bioeng. 2017, 124, 400–407. [Google Scholar] [CrossRef]
- Suzuki, T.; Seta, K.; Nishikawa, C.; Hara, E.; Shigeno, T.; Nakajima-Kambe, T. Improved ethanol tolerance and ethanol production from glycerol in a streptomycin-resistant Klebsiella variicola mutant obtained by ribosome engineering. Bioresour. Technol. 2015, 176, 156–162. [Google Scholar] [CrossRef]
- Ochi, K.; Hosaka, T. New strategies for drug discovery: Activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 2013, 97, 87–98. [Google Scholar] [CrossRef]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Genet. 2015, 13, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Okada, B.K.; Seyedsayamdost, M.R. Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 2017, 41, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Beppu, T. Antibiotics in microbial coculture. J. Antibiot. 2017, 70, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Sandiford, S.K. Genome database mining for the discovery of novel lantibiotics. Expert Opin. Drug Discov. 2017, 12, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-S. Phylogeny-guided (meta)genome mining approach for the targeted discovery of new microbial natural products. J. Ind. Microbiol. Biotechnol. 2016, 44, 285–293. [Google Scholar] [CrossRef]
- Hosaka, T.; Ohnishi-Kameyama, M.; Muramatsu, H.; Murakami, K.; Tsurumi, Y.; Kodani, S.; Yoshida, M.; Fujie, A.; Ochi, K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 2009, 27, 462–464. [Google Scholar] [CrossRef]
- Inaoka, T.; Takahashi, K.; Yada, H.; Yoshida, M.; Ochi, K. RNA polymerase mutation activates the production of a dormant antibiotic 3,3’-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 2004, 279, 3885–3892. [Google Scholar] [CrossRef]
- Derewacz, D.K.; Goodwin, C.R.; McNees, C.R.; McLean, J.A.; Bachmann, B.O. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 2336–2341. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Cui, C.-B.; Li, C.-W.; Hua, W.; Wu, C.-J.; Zhu, T.-J.; Gu, Q.-Q. Activation of dormant secondary metabolite production by introducing neomycin resistance into the deep-sea fungus, Aspergillus versicolor ZBY-3. Mar. Drugs 2014, 12, 4326–4352. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Jamison, M.; La, S.; Macmillan, J.B. Inducamides A–C, chlorinated alkaloids from an RNA polymerase mutant strain of Streptomyces sp. Org. Lett. 2014, 16, 5656–5659. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-J.; Yi, L.; Cui, C.-B.; Li, C.-W.; Wang, N.; Han, X. Activation of the silent secondary metabolite production by introducing neomycin-resistance in a marine-derived Penicillium purpurogenum G59. Mar. Drugs 2015, 13, 2465–2487. [Google Scholar] [CrossRef] [PubMed]
- Derewacz, D.K.; Covington, B.C.; McLean, J.A.; Bachmann, B.O. Mapping microbial response metabolomes for induced natural product discovery. ACS Chem. Boil. 2015, 10, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Thong, W.L.; Shin-Ya, K.; Nishiyama, M.; Kuzuyama, T. Methylbenzene-containing polyketides from a Streptomyces that spontaneously acquired rifampicin resistance: Structural elucidation and biosynthesis. J. Nat. Prod. 2016, 79, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Thong, W.L.; Shin-Ya, K.; Nishiyama, M.; Kuzuyama, T. Discovery of an antibacterial isoindolinone-containing tetracyclic polyketide by cryptic gene activation and characterization of its biosynthetic gene cluster. ACS Chem. Boil. 2018, 13, 2615–2622. [Google Scholar] [CrossRef]
- Qi, H.; Ma, Z.; Xue, Z.; Ruan, H.; Yu, X.; Gao, M.; Wang, J. Two new compounds from rifamycin resistant mutant strain Streptomyces sp. HS-NF-1046R. Acta. Pharm. Sin. B 2019, 54, 117–121. [Google Scholar]
- Jiménez, C. Marine natural products in medicinal chemistry. ACS Med. Chem. Lett. 2018, 9, 959–961. [Google Scholar] [CrossRef]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [Green Version]
- Cashel, M.; Gallant, J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 1969, 221, 838–841. [Google Scholar] [CrossRef]
- Lai, C.; Xu, J.; Tozawa, Y.; Okamoto-Hosoya, Y.; Yao, X.; Ochi, K. Genetic and physiological characterization of rpoB mutations that activate antibiotic production in Streptomyces lividans. Microbiology 2002, 148, 3365–3373. [Google Scholar] [CrossRef] [PubMed]
- Vigliotta, G.; Tredici, S.M.; Damiano, F.; Montinaro, M.R.; Pulimeno, R.; Di Summa, R.; Massardo, D.R.; Gnoni, G.V.; Alifano, P. Natural merodiploidy involving duplicated rpoB alleles affects secondary metabolism in a producer actinomycete. Mol. Microbiol. 2005, 55, 396–412. [Google Scholar] [CrossRef] [PubMed]
- Tala, A.; Wang, G.; Zemanova, M.; Okamoto, S.; Ochi, K.; Alifano, P. Activation of dormant bacterial genes by Nonomuraea sp. strain ATCC 39727 mutant-type RNA polymerase. J. Bacteriol. 2009, 191, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Artsimovitch, I.; Patlan, V.; Sekine, S.; Vassylyeva, M.; Hosaka, T.; Ochi, K.; Yokoyama, S.; Vassylyev, D.; Initiative, R.S.G. Structural basis for transcription regulation by alarmone ppGpp. Cell 2004, 117, 299–310. [Google Scholar] [CrossRef]
- Okamoto-Hosoya, Y.; Hosaka, T.; Ochi, K. An aberrant protein synthesis activity is linked with antibiotic overproduction in rpsL mutants of Streptomyces coelicolor A3(2). Microbiology 2003, 149, 3299–3309. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, T.; Xu, J.; Ochi, K. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol. Microbiol. 2006, 61, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Sato, S.; Tanaka, Y.; Ochi, K.; Hosaka, T. Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl. Environ. Microbiol. 2015, 81, 3869–3879. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Spiegelman, G.B.; Yim, G. The world of subinhibitory antibiotic concentrations. Curr. Opin. Microbiol. 2006, 9, 445–453. [Google Scholar] [CrossRef] [PubMed]



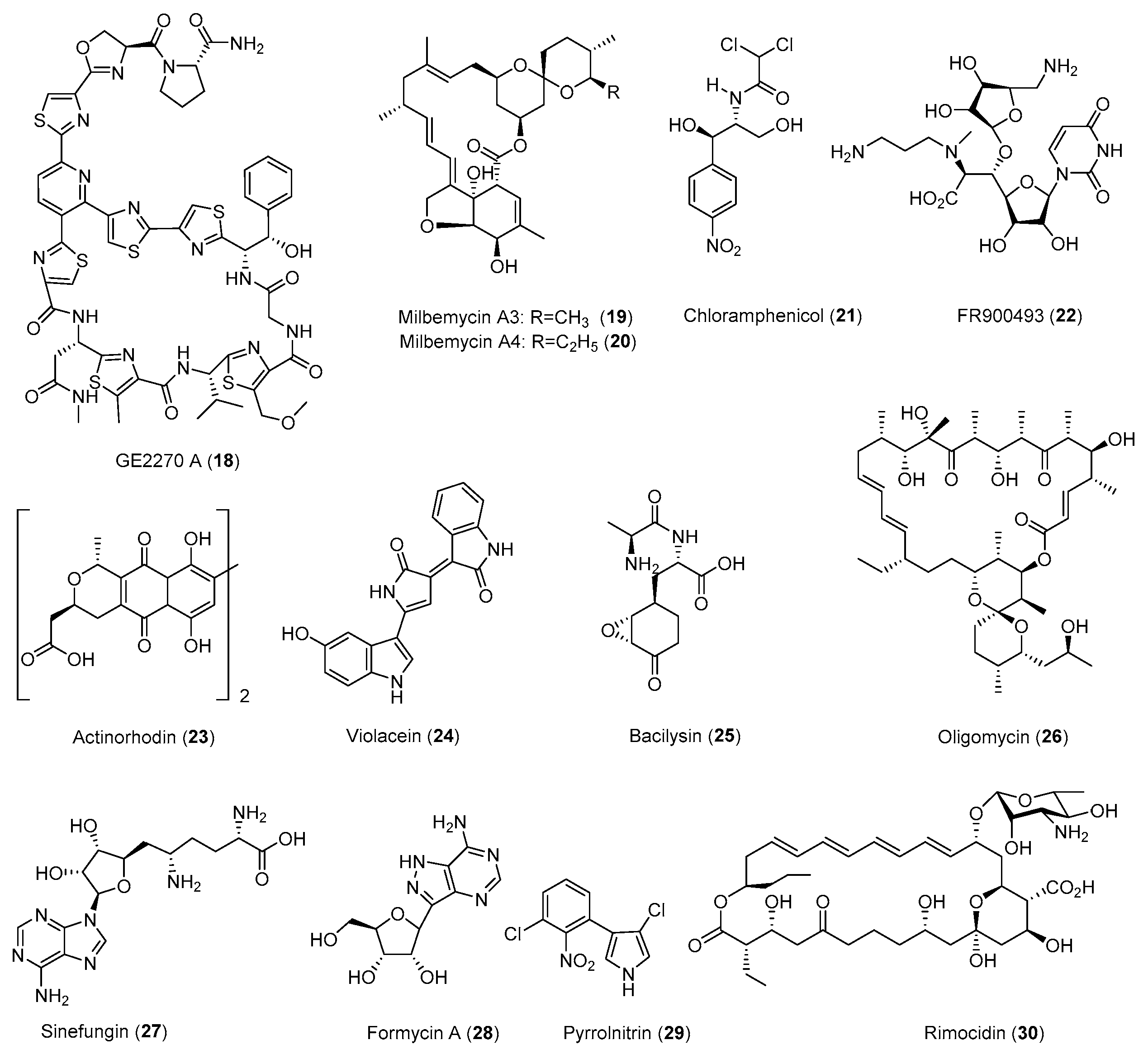
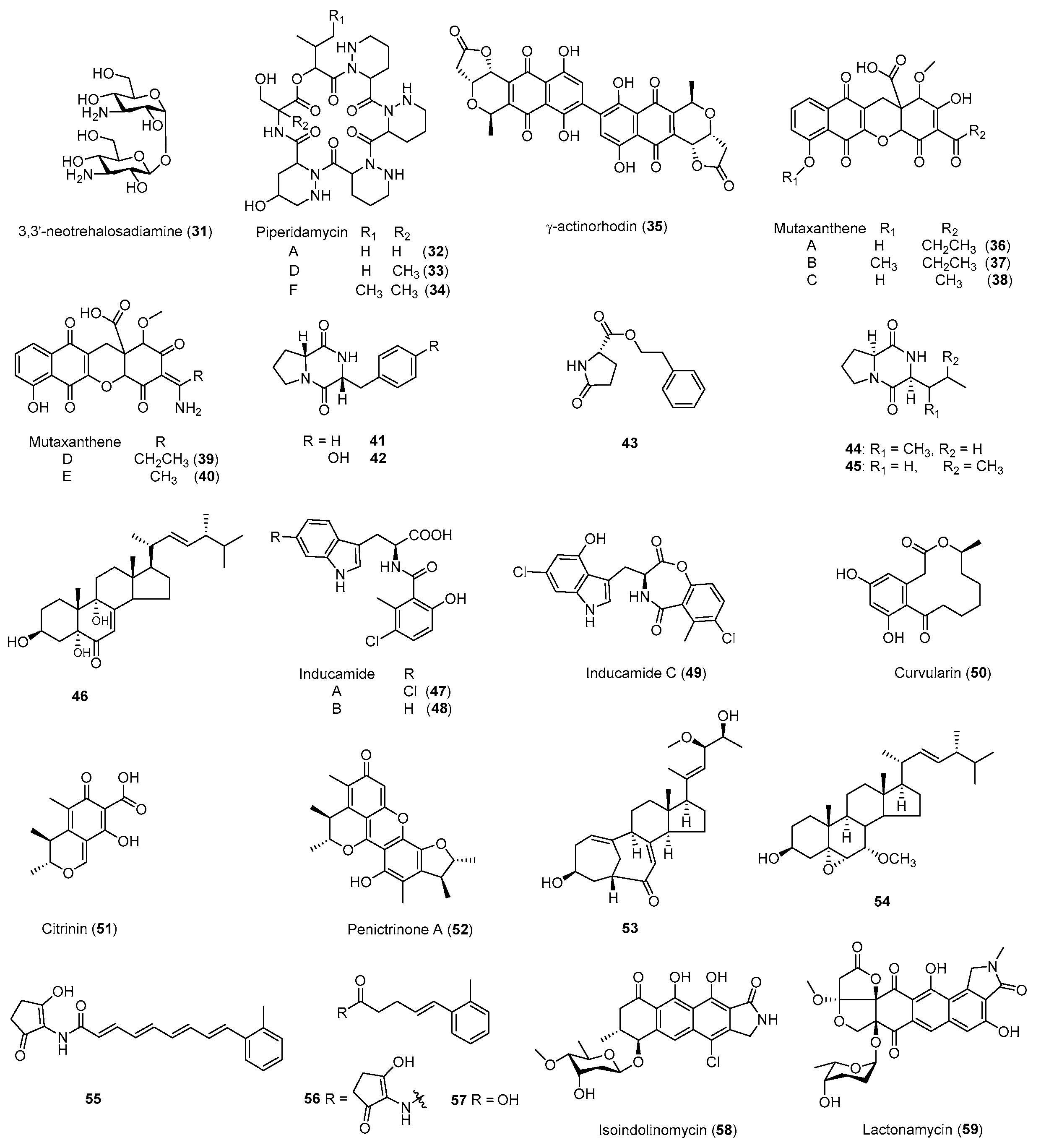
| Antibiotic | Strain | Method | Mutation a | (Fold)/(g/L) b | Year/Ref. |
|---|---|---|---|---|---|
| Actinorhodin (23) | S. coelicolor | Str, Tet | - c | - | 1996 [15] |
| S. coelicolor | Str | K88E | 15 (2.8 OD600) | 1997 [25] | |
| S. coelicolor | Par | P91S | 5–21 (2.1 OD600) | 2000 [26] | |
| S. coelicolor | Str, Gen, Rif | K88E | 48 (6.88 OD633) | 2001 [27] | |
| S. coelicolor | relA and relC mutants Rif | Q424L | >93 (2.79 OD633) | 2002 [28] | |
| S. coelicolor | Str, Rif, Par, Gen | - | 180 (1.63) | 2008 [29] | |
| S. coelicolor | Str | R86P | 55–106 (0.1338 ± 0.007) | 2009 [30] | |
| S. lividans | Ery | - | 6–8 (0.3) | 2012 [31] | |
| S. coelicolor | Rif | S433L | 42–55.5 (28.7 ± 1.3) | 2013 [32] | |
| Actinomycin D (9) | S. antibioticus | Str | - | 5.2 (0.063) | 1998 [33] |
| S.antibioticus | Gen | - | 4.1 (0.05) | 2008 [34] | |
| S. parvulus | Str | K88R | 2–10 (0.0328 ± 0.0086) | 2009 [30] | |
| S. antibioticus | Str | K88R | 7–10 (0.0471 ± 0.0044) | 2009 [30] | |
| S. parvulus | Rif | D427V | 1–2.2 (0.010 ± 0.001) | 2013 [32] | |
| S. antibioticus | Rif | H437R | 5–11 (0.086 ± 0.016) | 2013 [32] | |
| Avermectins (3,4) | S. avermitilis | frr overexpression | - | 3–3.7 (>0.8) | 2010 [35] |
| Avilamycin (5) | S. viridochromogenes | 60Co γ-ray, GS, Str | K43N | 36.8 (1.4) | 2013 [36] |
| Chloramphenicol (21) | S. coelicolor | Str, Rif, HE | - | 20–40 (−) | 2011 [37] |
| Congocidine | S. coelicolor | Str, Rif, HE | - | 20–40 (−) | 2011 [37] |
| Daptomycin (7) | S. roseosporus | Ple | - | 1.3 (>0.08) | 2013 [38] |
| S. roseosporus | Neo, Gen, Rif, Par, GS a | - | 4 (0.324) | 2018 [39] | |
| A21978C | S. roseosporus | Str, Reporter gene | K43N | 2.2 (>0.12) | 2012 [40] |
| Formycin A (28) | S. lavendulae | Str | - | 5.2 (0.13) | 1998 [33] |
| S. lavendulae | Str | R440H | 2.4–4.6 (0.055 ± 0.014) | 2013 [32] | |
| Fredericamycin A (15) | S. chattanoogensis | Str | - | 26 (0.26) | 1998 [33] |
| S. somaliensis | Rif | R444H | 3 (0.6795 ± 0.0158) | 2015 [41] | |
| Milbemycin (19,20) | S. bingchenggensis | CM, Str, UV | - | 1.8 (1.45) | 2009 [42] |
| Nosiheptide (1) | S. actuosus | 60Coγ-irradiation, LiCl, Str, GS | K88R | 9.2 (1.54) | 2014 [43] |
| Oligomycin (26) | S. avermitilis | Str | K43M | 20–40 (1.064) | 2009 [30] |
| Rimocidin (30) | S. rimosus | Gen, Rif | - | 2.5–6.2 (0.6731) | 2019 [44] |
| Salinomycin (6) | S. albus | Str, Gen, Rif | - | 2.3 (25) | 2003 [45] |
| Sinefungin (27) | S. incarnatus | Rif, L-Arg | D427G | 35 (>0.05) | 2010 [46] |
| Streptomycin (10) | S. griseus | Gen | - | 10 (0.3) | 2008 [34] |
| S. griseus | Rif | Q424K | 2.4–6.0 (0.178 ± 0.027) | 2013 [32] | |
| Tiancimycin A (17) | S. sp. CB03234 | Rif | L422P | 40 (0.0225 ± 0.0031) | 2016 [47] |
| 2018 [48] | |||||
| S. sp. CB03234 | Str | K43N | 45 (0.0137 ± 0.0003) | 2019 [49] | |
| Tiancimycins D (14) | S. sp. CB03234 | Str | K43N | 109 (0.0192 ± 0.0004) | 2019 [49] |
| Toyocamycin (16) | S. diastatochromogenes | frr overexpression | - | 1.46 (>0.6) | 2014 [50] |
| S. diastatochromogenes | Rif | H437Y | 4.5 (0.68) | 2016 [51] | |
| Virginiamycin (2) | S. virginiae | UV, GS, Str | - | 11.6 (0.251) | 2018 [52] |
| 6′-Deoxy-bleomycin Z (12) | S. flavoviridis | UV, Str, Gen, Rif | - | 7 (0.07) | 2018 [53] |
| Antibiotic | Strain | Method | Mutation a | (Fold)/(g/L) b | Year/Ref. |
|---|---|---|---|---|---|
| Bacilysin (25) | B. subtilis | Str, Rif | K56R L467P | 5–7 (0.0166 ± 0.0009) | 2015 [54] |
| Erythromycin (11) | S. erythraea | Rif | S444F | 4 (>1.5) | 2009 [55] |
| S. erythraead | Rif | H437R | 4.0 (0.163 ± 0.034) | 2013 [32] | |
| FR900493 (22) | B. cereus | Str | - c | 7.2 (0.55) | 1998 [33] |
| B. cereus | Gen | - | 2.7 (0.22) | 2008 [34] | |
| GE2270 A (18) | P. rosea | Gen, Str, Rif | - | 1.8 (−) | 2006 [56] |
| Norvancomycin | A. orientalis | Str, Rif, UV, HEE | - | 1.4 (−) | 2006 [57] |
| Pyrrolnitrin (29) | P. pyrrocinia | Str | - | 10 (0.015) | 1998 [33] |
| P.pyrrocinia | Gen | - | 5.3 (0.008) | 2008 [34] | |
| Vancomycin (8) | A. orientalis | Rif | S442Y | 2.6–3.4 (0.27 ± 0.017) | 2013 [32] |
| Violacein (24) | E. coli | Lin, Kan, HE | - | 41 (−) | 2011 [58] |
| Miscellaneous Products | Strain | Method | Mutation a | Fold/(g/L) b | Year/Ref. |
|---|---|---|---|---|---|
| CITase | P. agaridevorans | Str, Rif | K56R, R485H | 1100 (1104 ± 143 U/mL) | 2018 [60] |
| α-Amylase | B. subtilis | Str | K56R | 1.5 (4.0 U/mL) | 2006 [61] |
| Xylanase | S. viridochromogenes | Str | K88R | 1.14 (>60 U/mL) | 2013 [62] |
| Vitamin B12 | P. shermanii | Rif, Gen, Ery | H437Y, H447R | 5.2 (304 ± 3 µg/L/OD600) | 2017 [63] |
| ε-poly-L-Lysine | S. albulus | ARTP, Str, GS | - c | 1.71 (3.0) d | 2016 [64] |
| S. albulus | HT, Par | - | 1.45 (2.59) | 2017 [65] | |
| S. albulus | Str, Gen, Rif | K108R | 1.75–2.39 (3.83) | 2017 [66] | |
| S. albulus | Str | E85G | 1.79 (3.04) | 2019 [67] | |
| L-Isoleucine | Corynebacterium glutamicum | frr and fusA Overexpression | - | 1.76 (28.5) | 2015 [68] |
| Butanol | Clostridium Saccharoperbutylacetonicum | Str | K43N | 1.6 (16.5) | 2017 [69] |
| Ethanol | K. variicola | Str | K43N | 1.3 (34) | 2015 [70] |
| Natural Products | Strain | Method | Mutation a | Activity | Year/Ref. |
|---|---|---|---|---|---|
| Neotrehalosadiamine (31) | B. subtilis | Rif | S487L | Antibacterial | 2004 [80] |
| Piperidamycins (32–34) | S. sp. 631689 | Rif, Str, Gen | K88R | Antibacterial | 2009 [79] |
| γ-Actinorhodin (35) | S. coelicolor | Rif | - b | Antibacterial | 2013 [32] |
| Mutaxanthenes (36–40) | Nocardiaceae FU40ΔApoS8 | Rif, Str | - | - | 2013 [81] |
| 41–46 | Aspergillus versicolo ZBY-3 | Neo | - | Antitumor | 2014 [82] |
| Inducamides A-C (47–49) | S. sp. SNC-109 | Rif | X442F | - | 2014 [83] |
| 50–54 | Penicillium purpurogenum G59 | Neo, DMSO | - | Antibacterial Antitumor | 2015 [84] |
| Fredericamycin A (15) | S. somaliensis SCSIO ZH66 | Rif | R447H | Antitumor | 2015 [41] |
| 16 secondary metabolites | S. coelicolor | Rif, Str | - | Antibacterial | 2015 [85] |
| 55–57 | S. sp. SANK 60404 | Rif | H447D | - | 2016 [86] |
| 58, 59 | S. sp. SoC090715LN-16 | Rif | H447Y | Antibacterial | 2018 [87] |
| Cyclopentene derivatives | S. sp. HS-NF-1046R | Rif | - | - | 2019 [88] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, S.; Duan, Y.; Huang, Y. The Application of Ribosome Engineering to Natural Product Discovery and Yield Improvement in Streptomyces. Antibiotics 2019, 8, 133. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics8030133
Zhu S, Duan Y, Huang Y. The Application of Ribosome Engineering to Natural Product Discovery and Yield Improvement in Streptomyces. Antibiotics. 2019; 8(3):133. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics8030133
Chicago/Turabian StyleZhu, Saibin, Yanwen Duan, and Yong Huang. 2019. "The Application of Ribosome Engineering to Natural Product Discovery and Yield Improvement in Streptomyces" Antibiotics 8, no. 3: 133. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics8030133





