1. Introduction
Staphylococcus aureus is often found as a component of the human microbiota associated with skin, skin glands, and mucous membranes, particularly in the nose of healthy individuals [
1,
2]. In some cases,
S. aureus causes a wide range of soft human infections [
3], such as mild skin and soft tissue infections, as well as life-threatening pneumonia, bacteremia, osteomyelitis, endocarditis, sepsis, and toxic shock syndrome [
4], and it is implicated in both community-acquired and nosocomial infections [
2]. In addition to the infections listed above,
S. aureus is often responsible for scalded skin syndrome and staphylococcal foodborne diseases [
5,
6].
In addition to causing infections in humans,
S. aureus can also be the origin of infections in ruminants such as cattle, goats, and sheep, leading to clinical and subclinical mastitis. The pathogen spreads from the udder of the infected animal into raw milk, affecting the quality and quantity of milk and milk-derived products. Therefore, this pathogen represents a major economic problem for farmers and a serious problem for the dairy industry [
7].
S. aureus pathogenicity depends upon its capability to produce and secrete different toxins and virulence factors that contribute to colonization and invasion of the host and bacterial spread [
8]. The family of superantigen exotoxins is comprised of well-known secreted virulence factors, such as the staphylococcal enterotoxins (
se), the toxic shock syndrome toxin 1 (
tsst-1), and the exfoliative toxins (
eta and
etb). The latter are associated with staphylococcal scalded-skin syndrome [
5]. Until now, more than 20 staphylococcal enterotoxins that can cause food poisoning or enterotoxin-like proteins have been identified [
9].
In addition to the production of virulence factors,
S. aureus genome shows enormous plasticity with the consequent acquisition of transmissible genetic elements, coding for resistance proteins. One example is the
mecA gene that is present within the staphylococcal cassette chromosome
mec (SCC
mec) [
2]. The
mecA gene encodes an alternative penicillin binding protein, PBP2a [
1], that makes the bacterial strain resistant both to methicillin (MRSA) and all other β-lactam antibiotics [
2]. The ability to acquire horizontally resistant genes and the antibiotic pressure induces the emergence of multidrug-resistant (resistant to three or more classes of antibiotics)
S. aureus strains, which are considered a significant concern for public health.
Furthermore, it is well known that S. aureus produces biofilm, a thick extracellular exopolysaccharide layer which protects bacteria. Biofilm can be easily formed inside biomaterials as indwelling medical devices, often causing chronic diseases that are difficult to eradicate. Biofilm formation is a multifactorial event, controlled by quorum sensing and several proteins, such as the accessory gene regulator (Agr), the biofilm-associated protein (Bap), the intercellular adhesion protein (Ica), and the S. aureus surface protein (SasC).
More recently, antimicrobial resistance has been found even in previously unexplored environments, where antibiotic pressure is missing, and it has been demonstrated that food can serve as a vehicle for transmission of
S. aureus to the human population [
10,
11]. Indeed, the comparison between human isolates and animal-derived isolates was performed also in other studies [
12] suggesting that food, after handling and processing, could represent a source of human infection, and for food operators a source of food contamination.
The aim of this work was to compare the antibiotic resistance profile and biofilm production of S. aureus isolates derived from fourteen medical specimens and twenty-one animal-derived samples collected in Sicily. In addition to the phenotypic characterization, the isolates were compared for the presence of toxin genes and biofilm-related genes.
3. Discussion
In this study, we report phenotypic and molecular analysis carried out on human- and animal-derived
S. aureus isolates collected in Sicily. Our results demonstrate that
S. aureus isolates from human specimens were multi-resistant to antibiotics and produce more biofilm than the isolates collected from animal-derived samples. The high percentage (78.6%) of the human-derived isolates with multiple antibiotic resistance is in accordance with a recent study carried out in Serbia [
12]. On the other hand, in other studies the animal-derived isolates showed a higher biofilm production than human-derived isolates [
13]. The percentage of animal-derived isolates (47.6%) displaying multidrug-resistance was lower than those found in studies performed on other samples of animal origin. Indeed, in a study conducted on meat and dairy products collected in Puglia (Italy), 68.8% of the isolates were resistant to at least one antibiotic [
14], and in another study performed on meat and poultry in the United States, 52% of the isolates were multi-resistant [
15]. It is possible that the prevalence of an extensive and traditional farm management for ruminants in Sicily assures a lower circulation for multidrug-resistant clones, especially in healthy animals and in food derived from healthy animals, as in this study. In a previous study, in Ragusa Province in Sicily, on cows with mastitis reared in semi-intensive management, a higher prevalence of MRSA was detected [
16].
In our previous study [
17], resistance to penicillin was the most diffused in both human- and animal-derived isolates, even if a higher percentage (86%) was observed in human-derived isolates.
Besides penicillin, human-derived isolates showed a high prevalence of resistance to erythromycin (50%), similar to results obtained on isolates collected from patients with early postoperative orthopedic implant-based infections [
18] (erythromycin 82%) and from swine, farmers, and abattoir workers [
19] (penicillin 96%, erythromycin 80.7%), even if the percentage of antibiotic resistant in our isolates was lower. In addition, we found less MRSA (35.7%) with respect to a study conducted on
S. aureus isolates from skin and soft tissue infection, bloodstream infection, and lower respiratory tract infection collected from South Italy (40.7%) [
20]. By comparing our results with this latter study, we found an increase of resistance to clindamycin (42.8 vs. 33%) oxacillin (36 vs. 0.9%), tetracycline (21.4 vs. 12.6%), and sulfamethoxazole-trimethoprim (7.1 vs. 3.2%) and a decrease of erythromycin (50 vs. 65%), moxifloxacin (11 vs. 72.3%), and gentamicin (21.4 vs. 39.5%) resistant isolates. A similar percentage of vancomycin, linezolid, and tigecycline resistant isolates was obtained.
With regards to animal-derived isolates, most strains showed resistance to penicillin (52%) and tetracycline (33%) in according with previous works [
11,
17,
20,
21,
22,
23]. By comparing our results with those obtained in a previous study [
6] which reported the antimicrobial profile of 80 isolates collected between 1998–2014, we found an increase of resistance to penicillin (52% vs. 35.7%), tetracycline (33% vs. 20%), ceftriaxone (14% vs. 3.7%), and gentamicin (14% vs. 4%), which suggested the spread of resistance. In contrast, the percentage of lincomycin resistant isolates (4.7% vs. 3.7%) was unchanged and erythromycin resistance was lower in the new isolates (0 vs. 2.5%). We cannot rule out that the low number of isolates can be a bias for this result. Moreover, we found that 43% of the animal-derived isolates had an intermediate susceptibility to erythromycin, suggesting that this antibiotic should be used in a controlled manner even in veterinary practice.
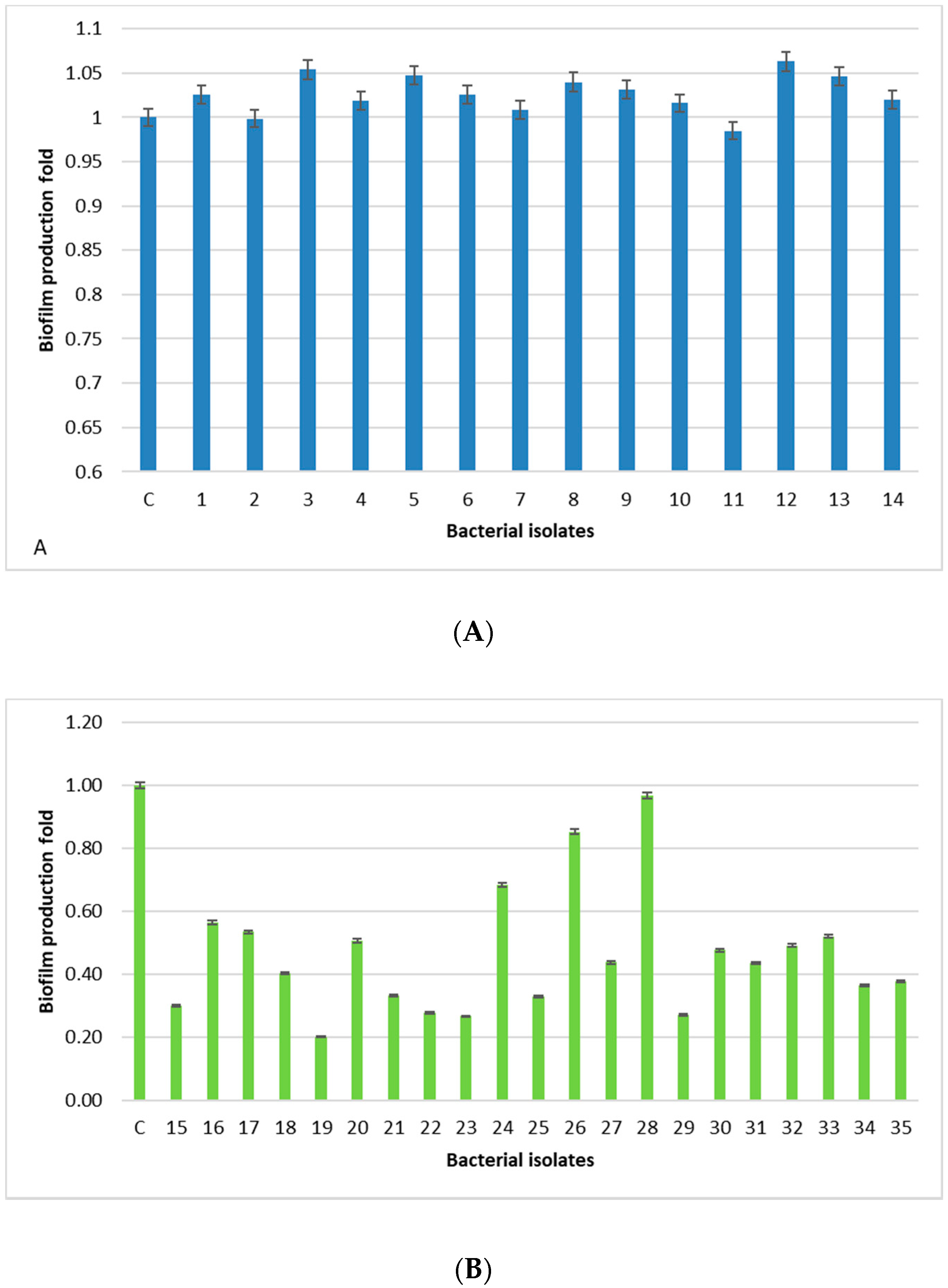
All human-derived isolates are biofilm formers, with the isolates 3, 5, 12, and 13 being the strongest, whereas, isolates derived from food, dairy products, and animal tissue samples have a weak/moderate biofilm capability, except for one isolate (28) showing a level comparable to the positive control. The lowest prevalence of the
bap PCR positive animal-derived samples (9.5%) is in accordance with the low ability of these isolates to produce biofilm, since
bap could facilitate biofilm production in mastitis [
24]. Moreover, the higher percentage of animal samples (76.2%) containing the
ica locus, with respect to the 14.2% of human-derived isolates, is in accordance with a study carried out in Iran [
13]. In this study,
sasC gene is involved, but not essential, in the biofilm formation process in
S. aureus in accordance with Schroeder [
25]. In fact, we found this gene in eight human-derived isolates (57%) and eight isolates derived from food, dairy products, and animal tissue samples (38%).
Antimicrobial spreading is a significant concern for public health worldwide. We analyzed the human- and animal-derived isolates for antibiotic resistance and virulence factors. The presence of the mecA gene was quite diffused (36%) in human-derived isolates and very low in animal-derived isolates (5%).
Striking differences between human- and animal-derived isolates were also found in the relative presence of enterotoxin genes. Interestingly, in human-derived isolates the simultaneous presence of the enterotoxin genes,
seg and
sei, was quite frequent (50%). In our previous study, only a single animal-derived isolate and four human-derived isolates derived from a severe poisoning case showed the simultaneous presence of
seg and
sei [
6]. In another recent study,
sei was the second most diffuse enterotoxin in humans [
12]. In addition, the animal strains showed the simultaneous presence of
sea and
see at 14.3%, according to another study on enterotoxin-producing
S. aureus isolated from mastitic cows [
26].
It is possible that the main reason for these differences is that human isolates are derived from symptomatic non-hospitalized patients subjected to microbiological controls while those derived from animals are part of a systematic screening. However, the results of this study confirm the importance of controlling antibiotic use in medical and veterinary practice. Although the S. aureus human-derived isolates could be more virulent for their antibiotic resistance, biofilm production, and presence of virulence genes, our results suggest monitoring the animal-derived isolates, since they are developing a greater resistance to the most commonly used antibiotics.