Antibiotics in Food Chain: The Consequences for Antibiotic Resistance
Abstract
:1. Introduction and Background
2. Drug Resistance Continuum
3. The Detrimental Effects of Antibiotics Misuse
4. Livestock as a Major Contributor of Antibiotic Resistance
| Sl. No. | Bacterial Species | Infection | Antibiotic Resistance Pattern | Sources of Human Infection | Genes |
|---|---|---|---|---|---|
| 1 | Campylobacter spp. | Gastrointestinal sequelae: Guillain-Barré syndrome | Fluoroquinolones, erythromycin | Food-producing animals (poultry) | tetO, gyrA [39,40] |
| 2 | Enterococcus spp. | Sepsis, urinary tract | Aminoglycosides ampicillin vancomycin | Food-producing animals (poultry); People exposed to hospital care, food animals | Tuf, VanC-1, VanC-2-VanC-3, pbp5 [41,42,43,44,45] |
| 3 | E. coli | Gastrointestinal, urinary tract, diarrhoea | Quinolones sulphonamides trimethoprim | Childcare facilities | Bla, qnrS, frdD [46,47,48] |
| 4 | Salmonella spp. (non-typhoidal) | Gastrointestinal, diarrhoea | Cephalosporins quinolones tetracyclines | Food-producing animals (pigs, cows, poultry) | IntI1, qnrA [49,50,51,52] |
| 5 | S. pneumoniae | Otitis media, pneumonia, sinusitis, meningitis | Penicillin, macrolides, cephalosporins, tetracyclines | Childcare facilities, paediatric populations | erm(B), mef [53,54,55,56] |
| 6 | S. pyogenes | Pharyngitis, impetigo, cellulitis | Macrolides, tetracyclines | Childcare facilities, paediatric Populations, schools | ermB, ermA and mefA [57] |
| 7 | S. aureus | ||||
| Community-associated | Skin, soft tissue, pneumonia, sepsis | Methicillin, cephalosporins, macrolides | Childcare facilities, injections, drug users | erm(A), erm(C), tetK, tetM, aacA-aphD, vat(A), vat(B) and vat(C) [58,59] | |
| Healthcare-associated | Endocarditis, pneumonia, sepsis | Methicillin, cephalosporins, quinolones, aminoglycosides, macrolides | People exposed to healthcare facilities such as nursing homes, dialysis, recent surgery or hospitalization | ||
| 8 | N. gonorrhoeae | Urethritis, pelvic inflammatory disease | Penicillin, cephalosporins, quinolones | Commercial sex workers | penA, penB, NorM [60,61] |
5. Scale of Antibiotic Use in Animals and Humans
6. Anthropogenic Contamination of Environment with Antibiotics and ARGs
7. Alternative Strategies to Combat Antibiotic Resistance
7.1. Phage or Bacteriophage Therapies
7.2. Predatory Bacteria
| Causative Agent | Model/Route | Condition | Type of Phage | Result |
|---|---|---|---|---|
| Shigella dysenteriae | Human/Oral | Dysentery | Cholera Bacteriophage | Recovered after 24 h [92] |
| P. aeruginosa | Murine/Oral | Sepsis | Phage strain KPP10 | 66.7% mortality reduction [93] |
| Vancomycin-resistant E. faecium | Murine/Intraperitoneal injection (i.p.) | Bacteremia | Phage strain C33 & ENB6 | 100% mortality reduction [94] |
| C. difficile | Hamster/Oral | Lleocecitis | Phage strain 135I | Prevented infection [95] |
| Vibrio cholera | Human/Oral | Cholera | Cholera Bacteriophage | 93% survival in treated group vs. 37% in control group [92] |
| Imipenem-resistant P. aeruginosa | Murine/I. p. injection | Bacteremia | Phage strain Ø9882 | 100% mortality reduction [96] |
| B-lactamase producing E. coli | Murine/I. p. injection | Bacteremia | Phage strain Ø9882 | 100% mortality reduction [96] |
| S. aureus | Rabbit/Subcutaneous injection | Wound Infection | Phage LS2a | Prevented infection [97] |
| Salmonella Typhi | Human/Oral | Typhoid | Pyophage, Intestiphage, Staphylococcal bacteriophage, PhageBioDerm | 5 fold decrease in typhoid incidence compared to placebo [98] |
| MDR S. aureus | Human/Tropical | Diabetic foot ulcer | Staphylococcal Phage Sb-1 | 100% recovery [99] |
| Antibiotic-resistant P. aeruginosa | Human/Oral | Chronic otitis | Biophage-PA | Improved symptoms in double-blind, placebo-controlled phase I/II trial [85] |
| E. coli | Murine/I. p. or subcutaneous injection | Meningitis and sepsis | Lytic Phage EC200PP | 100% and 50% mortality reduction meningitis and sepsis, respectively [100] |
7.3. Immunotherapeutics
7.4. Haemofiltration Devices
7.5. Quorum-Sensing Inhibitors
- (1)
- (2)
- Furanosyl borate (Autoinducer-2, AI-2),
- (3)
- N-acyl homoserine lactones (AHLs),
- (4)
- Methyl-dodecanoic acid, and
- (5)
7.6. Antimicrobial Adjuvants (AA)
- (a)
- Biofilm disruption.
- (b)
- Augmenting the uptake of antimicrobial in the target cell.
- (c)
- Enhancing the oxidative stress in bacteria.
- (d)
- Supressing the ARG.
- (e)
- Inhibition of bacterial efflux pumps.
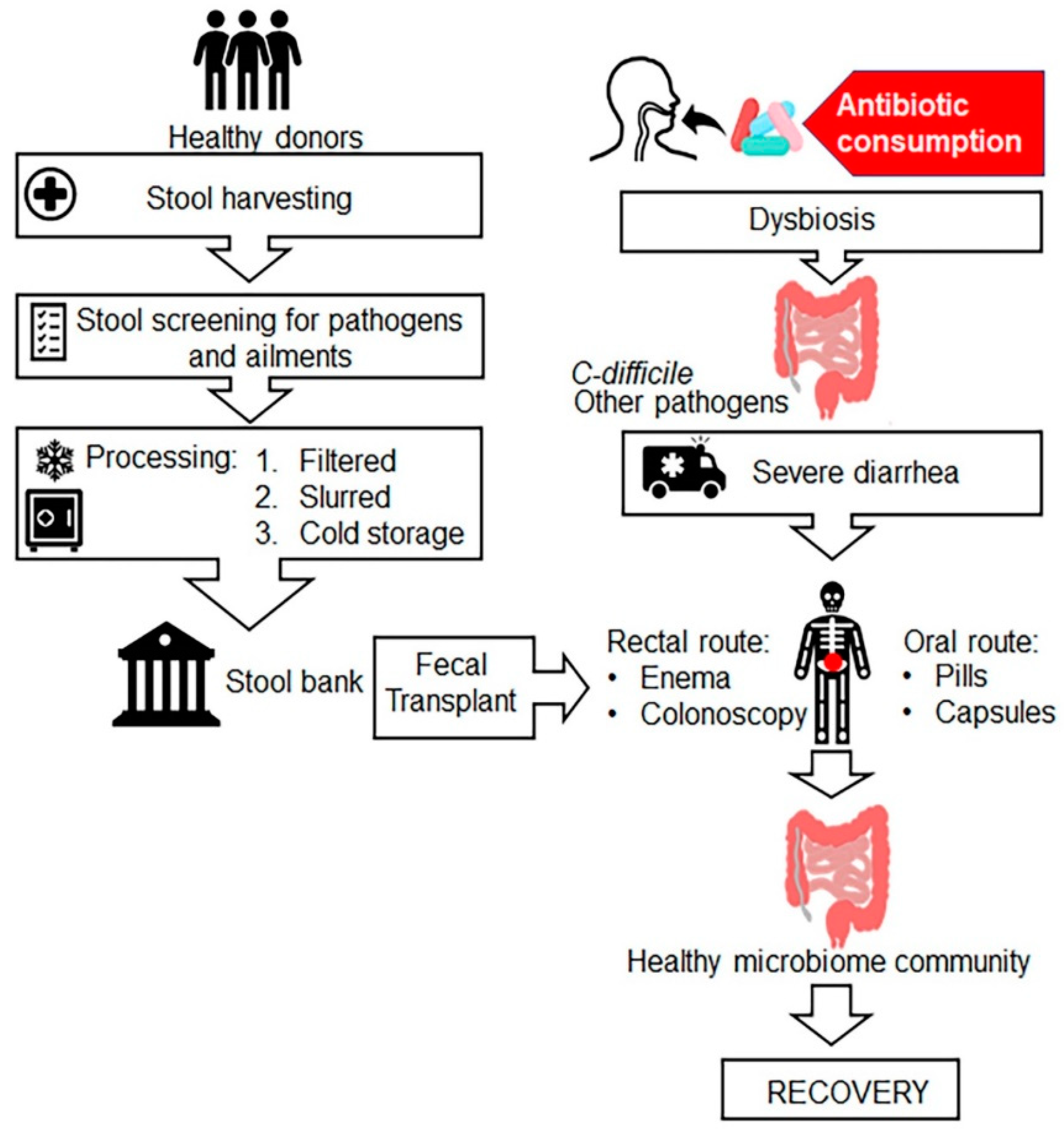
7.7. Faecal Microbiota Transplantation (FMT)
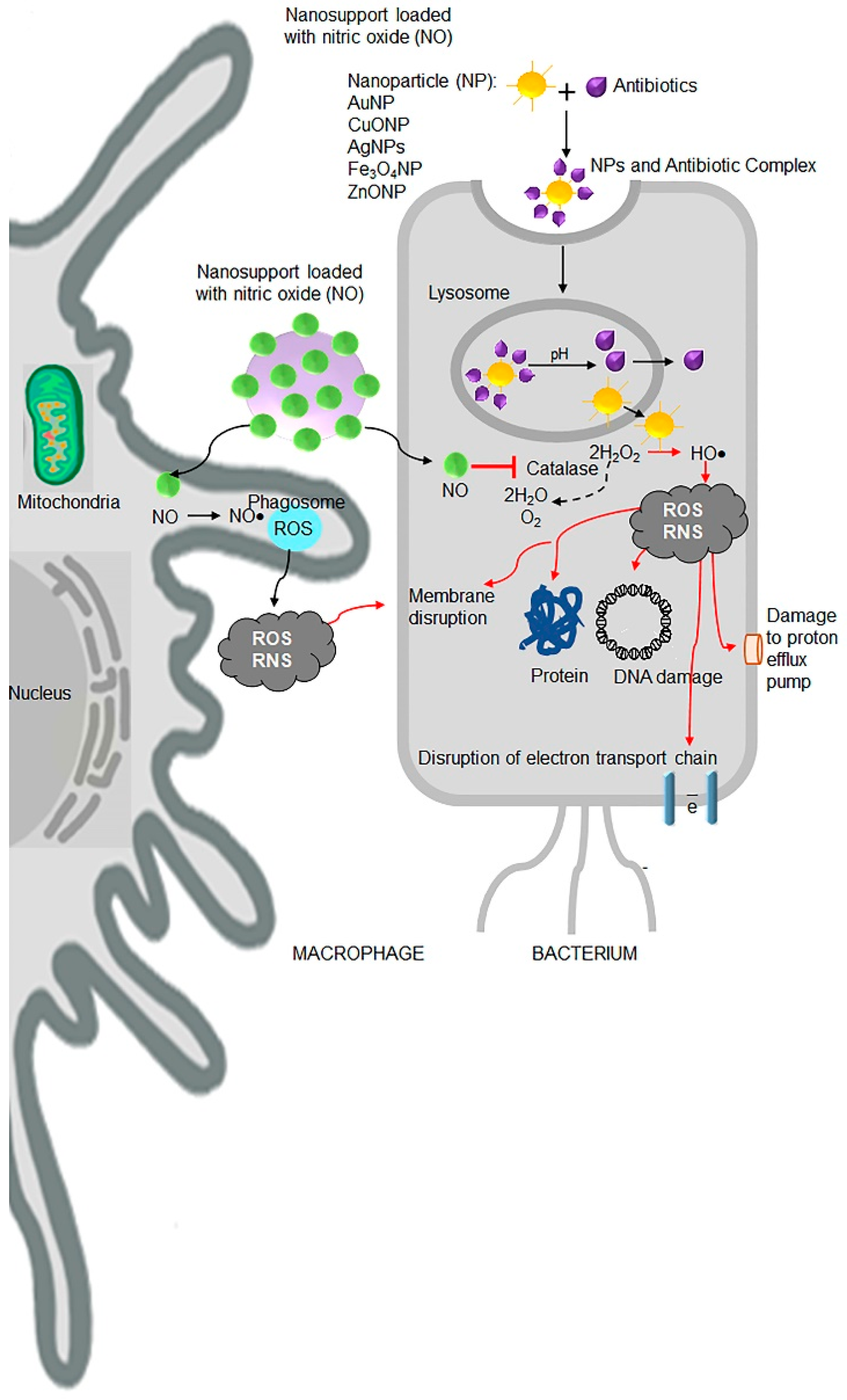
7.8. Nanoantibiotics
| S.No. | Metal Nanoparticles Used | Action against Bacteria |
| 1 | Silver | E. coli, M. tuberculosis, MRSA, S. aureus, S. pyogens, K. pneumonia, [166,167,168,169,170,171,172,173,174,175]. |
| 2 | Titanium | K. pneumonia, S. aureus, A. baumannii, E. coli, Morganella morganii [152] |
| 3 | Gold | MRSA, E. coli, P. aeruginosa, S. aureus, Enterococcus spp., B. subtilis [151,176,177] |
| S.No. | Metal Oxide Nanoparticles Used | Action against Bacteria |
| 1 | Zinc oxide | MRSA, Streptococcus agalactiae [155] |
| 2 | Manganese oxide | MRSA [178] |
| 3 | Manganese oxide | E. coli [156] |
| S.No. | Metal and Metal Oxide Nanoparticle Composite Used | Action against Bacteria |
| 1 | Zinc doped copper oxide nanocomposite | MRSA, E. coli [154] |
| 2 | Copper doped zinc oxide nanocomposite | E. coli, S. aureus [179] |
| S.No. | Metal Oxide Nanoparticles in Combination with Antibiotics Used | Action against Bacteria |
| 1 | ZnO and Antibiotics (cefotaxime, ampicillin, ceftriaxone, and cefepime) | E. coli, K. pneumoniae, Sphingomonas paucimobilis, and P. aeruginosa, respectively [180] |
| 2 | TiO2 nanoparticles in combination with antibiotics (β-lactams, cephalosporin, glycopeptides, aminoglycosides, flouroquinolones, azlides, macrolides, lincosamides, and sulphonamides) | Showed improved antibacterial activity [181] |
| S.No. | Metal Nanoparticles in Combination with Antibiotics | Action against Bacteria |
| 1 | Gold nanoparticles and Ampicillin | MDR P. aeruginosa, E. aerogenes, and MRSA [182] |
| 2 | AgNPs with ciprofloxacin, imipenem, gentamycin, trimethoprim, and vancomycin | MDR E. coli, P. aeruginosa, E. faecalis, S. aureus, Micrococcus luteus, A. baumannii, K. pneumoniae, and Bacillus spp. [183] |
7.9. Plant-Derived Antimicrobials and Essential Oils
7.10. Probiotics, Postbiotics and Synbiotics
| S.No. | Essential Oils (Components) | Active against Bacteria |
|---|---|---|
| 1 | Mentha (menthol, isomenthone, limonene, iso-menthanol, menthol acetate, carvone, β-pinene, α-pinene, 1,8-cineole, α-terpineol, isopulegol, pulegone, piperiton, piperitone oxide, and β-phellandrene.) | S. aureus, Staphylococcus epidermidis, B. cereus, and E. coli, S. pyogenes, P. aeruginosa, Pseudomonas fluorescens, C. albicans, and V. cholerae, [200,201,202] |
| 2 | Basil (Linalool, epi-α-cadinol, α-bergamotene, γ-cadinene, germacrene D, camphor. methylchavikol, methylcinnamat, linolen, eugenol, cis-geraniol, 1,8-cineole, α-bergamotene, β-caryophyllene, viridiflorol.) | S. aureus and B. subtilis, Staphylococcus, Pseudomonas, and Enterococcus genera, L. monocytogenes and B. cereus Vibrio spp. and Aerobacter hydrophila [203,204,205] |
| 3 | Oregano (thymol, carvacrol, ρ-cymene, thymoquinone, and γ-terpinene.) | Sarcina lutea, S. aureus, C. albicans, E. faecalis, and B. cereus [206,207] |
| 4 | Rosemary (α-pinene, myrcene, 1,8-cineole, camphor, camphene, α-terpineol, and borneol.) | S. epidermidis, S. aureus, B. subtilis, Proteus vulgaris, P. aeruginosa, and E. coli. [208,209,210,211] |
7.11. RNA Therapy
7.12. Development and Use of Vaccines
8. Mitigation Steps to Curb the Menace of Antibiotic Resistance
- Strengthening of surveillance data.
- Improving awareness of antibiotic resistance.
- Improving the practices of antibiotic prescription.
- Improvement of poor sanitation, malnutrition, and endemic infections.
- Optimizing the use of antimicrobial medicines and restricting over the counter sale of antibiotics.
- Improving the public awareness and government commitment.
- Reducing the incidence of infection by various means.
- Reducing clinical trial risk.
- Boosting market value for not feeding animals antibiotics.
- Strengthening the regulation of farm feeding of antibiotics.
- Ensuring the quality of generic antibiotics.
- Early sharing of data.
- Organizing world antibiotic awareness week.
- Implementation of the global antimicrobial resistance surveillance system (GLASS).
- Establishing the global antibiotic research and development partnership (GARDP).
- Establishing the interagency coordination group on antimicrobial resistance (IACG).
9. Future Strategies, Challenges, and Outlooks
Author Contributions
Funding
Conflicts of Interest
References
- Tan, S.Y.; Tatsumura, Y. Alexander Fleming (1881–1955): Discoverer of penicillin. Singap. Med. J. 2015, 56, 366–367. [Google Scholar] [CrossRef] [Green Version]
- Hayden, G.E.; Tuuri, R.E.; Scott, R.; Losek, J.D.; Blackshaw, A.M.; Schoenling, A.J.; Nietert, P.J.; Hall, G.A. Triage sepsis alert and sepsis protocol lower times to fluids and antibiotics in the ED. Am. J. Emerg. Med. 2016, 34, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- The Bacterial Challenge: Time to React; ECDC/EMEA Joint Technical Report; European Medicines Agency and European center for Disease Control: Stockholm, Sweden, 2009. [CrossRef]
- Holmberg, S.D.; Solomon, S.L.; Blake, P.A. Health and economic impacts of antimicrobial resistance in Thailand. J. Health Serv. Res. Policy 2012, 6, 352–360. [Google Scholar]
- Available online: https://www.cdc.gov/drugresistance/ (accessed on 1 August 2020).
- de Kraker, M.E.; Stewardson, A.J.; Harbarth, S. Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef] [Green Version]
- Xie, R.; Zhang, X.D.; Zhao, Q.; Peng, B.; Zheng, J. Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg. Microbes Infect. 2018, 7, 31. [Google Scholar] [CrossRef] [Green Version]
- Laxminarayan, R.; Chaudhury, R.R. Antibiotic Resistance in India: Drivers and Opportunities for Action. PLoS Med. 2016, 13, e1001974. [Google Scholar] [CrossRef] [Green Version]
- Ganguly, N.K.; Arora, N.K.; Chandy, S.J.; Fairoze, M.N.; Gill, J.P.; Gupta, U.; Hossain, S.; Joglekar, S.; Joshi, P.C.; Kakkar, M.; et al. Rationalizing antibiotic use to limit antibiotic resistance in India. Indian J. Med. Res. 2011, 134, 281–294. [Google Scholar]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef]
- Armstrong, G.L.; Conn, L.A.; Pinner, R.W. Trends in infectious disease mortality in the United States during the 20th century. JAMA 1999, 281, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Yadav, A.K.; Verma, V.; Singh, B.; Mal, G.; Nagpal, R.; Hemalatha, R. Bioengineered probiotics as a new hope for health and diseases: An overview of potential and prospects. Future Microbiol. 2016, 11, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaman, S.B.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Sommer, M.O.A.; Munck, C.; Toft-Kehler, R.V.; Andersson, D.I. Prediction of antibiotic resistance: Time for a new preclinical paradigm? Nat. Rev. Microbiol. 2017, 15, 689–696. [Google Scholar] [CrossRef] [Green Version]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [Green Version]
- Planta, M.B. The role of poverty in antimicrobial resistance. J. Am. Board Fam. Med. 2007, 20, 533–539. [Google Scholar] [CrossRef]
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Hall, B.G.; Salipante, S.J.; Barlow, M. Independent origins of subgroup Bl + B2 and subgroup B3 metallo-beta-lactamases. J. Mol. Evol. 2004, 59, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701. [Google Scholar] [CrossRef]
- Wright, G.D. Q&A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 123. [Google Scholar] [CrossRef] [Green Version]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Marchant, J. When antibiotics turn toxic. Nature 2018, 555, 431–433. [Google Scholar] [CrossRef] [Green Version]
- Allen, H.K. Antibiotic resistance gene discovery in food-producing animals. Curr. Opin. Microbiol. 2014, 19, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kakkar, M.; Walia, K.; Vong, S.; Chatterjee, P.; Sharma, A. Antibiotic resistance and its containment in India. BMJ 2017, 358, j2687. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [Green Version]
- Food and Drug Administration (FDA). Antimicrobials Sold or Distributed for Use in Food-Producing Animals; US Food and Drug Administration: Silver Spring, MD, USA, 2015.
- Furuya, E.Y.; Lowy, F.D. Antimicrobial-resistant bacteria in the community setting. Nat. Rev. Microbiol. 2006, 4, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [Green Version]
- General Accounting Office (US). Antibiotic Resistance: Federal Agencies Need to Better Focus Efforts to Address Risk to Humans from Antibiotic Use in Animals. Report to Congressional Requesters. April 2004. Available online: http://www.gao.gov/new.items/d04490.pdf (accessed on 1 August 2020).
- Hawkey, P.M. The growing burden of antimicrobial resistance. J. Antimicrob. Chemother. 2008, 62 (Suppl. s1), i1–i9. [Google Scholar] [CrossRef]
- Martínez, J.L. Natural antibiotic resistance and contamination by antibiotic resistance determinants: The two ages in the evolution of resistance to antimicrobials. Front. Microbiol. 2012, 3, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iovine, N.M. Resistance mechanisms in Campylobacter jejuni. Virulence 2013, 4, 230–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.E.; Besser, J.M.; Hedberg, C.W.; Leano, F.T.; Bender, J.B.; Wicklund, J.H.; Johnson, B.P.; Moore, K.A.; Osterholm, M.T. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. Investigation Team. N. Engl. J. Med. 1999, 340, 1525–1532. [Google Scholar] [CrossRef]
- Bates, J.; Jordens, J.Z.; Griffiths, D.T. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J. Antimicrob. Chemother. 1994, 34, 507–514. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef]
- Morris, J.G., Jr.; Shay, D.K.; Hebden, J.N.; McCarter, R.J., Jr.; Perdue, B.E.; Jarvis, W.; Johnson, J.A.; Dowling, T.C.; Polish, L.B.; Schwalbe, R.S. Enterococci resistant to multiple antimicrobial agents, including vancomycin. Establishment of endemicity in a university medical center. Ann. Intern. Med. 1995, 123, 250–259. [Google Scholar] [CrossRef]
- Pillay, S.; Zishiri, O.T.; Adeleke, M.A. Prevalence of virulence genes in Enterococcus species isolated from companion animals and livestock. Onderstepoort J. Vet. Res. 2018, 85, e1–e8. [Google Scholar] [CrossRef] [Green Version]
- Clark, N.C.; Teixeira, L.M.; Facklam, R.R.; Tenover, F.C. Detection and differentiation of vanC-1, vanC-2, and vanC-3 glycopeptide resistance genes in enterococci. J. Clin. Microbiol. 1998, 36, 2294–2297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sáenz, Y.; Briñas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother. 2004, 48, 3996–4001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Liu, Q.; Teng, Y.; Ou, L.; Xi, Y.; Chen, S.; Duan, G. The resistance mechanism of Escherichia coli induced by ampicillin in laboratory. Infect. Drug Resist. 2019, 12, 2853–2863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornasini, M.; Reves, R.R.; Murray, B.E.; Morrow, A.L.; Pickering, L.K. Trimethoprim-resistant Escherichia coli in households of children attending day care centers. J. Infect. Dis. 1992, 166, 326–330. [Google Scholar] [CrossRef]
- Gunell, M.; Webber, M.A.; Kotilainen, P.; Lilly, A.J.; Caddick, J.M.; Jalava, J.; Huovinen, P.; Siitonen, A.; Hakanen, A.J.; Piddock, L.J. Mechanisms of resistance in nontyphoidal Salmonella enterica strains exhibiting a nonclassical quinolone resistance phenotype. Antimicrob. Agents Chemother. 2009, 53, 3832–3836. [Google Scholar] [CrossRef] [Green Version]
- Alam, S.B.; Mahmud, M.; Akter, R.; Hasan, M.; Sobur, A.; Nazir, K.N.H.; Noreddin, A.; Rahman, T.; El Zowalaty, M.E.; Rahman, M. Molecular Detection of Multidrug Resistant Salmonella Species Isolated from Broiler Farm in Bangladesh. Pathogens 2020, 9, 201. [Google Scholar] [CrossRef] [Green Version]
- Mølbak, K.; Gerner-Smidt, P.; Wegener, H.C. Increasing quinolone resistance in Salmonella enterica serotype Enteritidis. Emerg. Infect. Dis. 2002, 8, 514–515. [Google Scholar] [CrossRef]
- Chiu, C.H.; Wu, T.L.; Su, L.H.; Chu, C.; Chia, J.H.; Kuo, A.J.; Chien, M.S.; Lin, T.Y. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype choleraesuis. N. Engl. J. Med. 2002, 346, 413–419. [Google Scholar] [CrossRef]
- Kresken, M.; Henrichfreise, B.; Bagel, S.; Brauers, J.; Wiedemann, B. High prevalence of the ermB gene among erythromycin-resistant streptococcus pneumoniae isolates in Germany during the winter of 2000-2001 and in vitro activity of telithromycin. Antimicrob. Agents Chemother. 2004, 48, 3193–3195. [Google Scholar] [CrossRef] [Green Version]
- Cherazard, R.; Epstein, M.; Doan, T.L.; Salim, T.; Bharti, S.; Smith, M.A. Antimicrobial Resistant Streptococcus pneumoniae: Prevalence, Mechanisms, and Clinical Implications. Am. J. Ther. 2017, 24, e361–e369. [Google Scholar] [CrossRef]
- Jacobs, M.R.; Bajaksouzian, S.; Zilles, A.; Lin, G.; Pankuch, G.A.; Appelbaum, P.C. Susceptibilities of Streptococcus pneumoniae and Haemophilus influenzae to 10 oral antimicrobial agents based on pharmacodynamic parameters: 1997 U.S. Surveillance study. Antimicrob. Agents Chemother. 1999, 43, 1901–1908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neto, A.S.; Lavado, P.; Flores, P.; Dias, R.; Pessanha, M.A.; Sousa, E.; Palminha, J.M.; Caniça, M.; Esperança-Pina, J. Risk factors for the nasopharyngeal carriage of respiratory pathogens by Portuguese children: Phenotype and antimicrobial susceptibility of Haemophilus influenzae and Streptococcus pneumoniae. Microb. Drug Resist. 2003, 9, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Bingen, E.; Bidet, P.; Mihaila-Amrouche, L.; Doit, C.; Forcet, S.; Brahimi, N.; Bouvet, A.; Cohen, R. Emergence of macrolide-resistant Streptococcus pyogenes strains in French children. Antimicrob. Agents Chemother. 2004, 48, 3559–3562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strommenger, B.; Kettlitz, C.; Werner, G.; Witte, W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 4089–4094. [Google Scholar] [CrossRef] [Green Version]
- Adwan, G.; Adwan, K.; Jarrar, N.; Amleh, A. Molecular detection of nine antibiotic resistance genes in methicillin resistant Staphylococcus aureus isolates. Roum. Arch. Microbiol. Immunol. 2014, 73, 9–17. [Google Scholar]
- Demczuk, W.; Sidhu, S.; Unemo, M.; Whiley, D.M.; Allen, V.G.; Dillon, J.R.; Cole, M.; Seah, C.; Trembizki, E.; Trees, D.L.; et al. Neisseria gonorrhoeae Sequence Typing for Antimicrobial Resistance, a Novel Antimicrobial Resistance Multilocus Typing Scheme for Tracking Global Dissemination of N. gonorrhoeae Strains. J. Clin. Microbiol. 2017, 55, 1454–1468. [Google Scholar] [CrossRef] [Green Version]
- Bhuiyan, B.U.; Rahman, M.; Miah, M.R.; Nahar, S.; Islam, N.; Ahmed, M.; Rahman, K.M.; Albert, M.J. Antimicrobial susceptibilities and plasmid contents of Neisseria gonorrhoeae isolates from commercial sex workers in Dhaka, Bangladesh: Emergence of high-level resistance to ciprofloxacin. J. Clin. Microbiol. 1999, 37, 1130–1136. [Google Scholar] [CrossRef] [Green Version]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.A.; Klugman, K.; Davies, S. Access to effective antimicrobials: A worldwide challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Bank. Available online: www.worldbank.org (accessed on 10 March 2015).
- Scholz, M.; Lee, B.-H. Constructed wetlands: A review. Int. J. Environ. Stud. 2005, 62, 421–447. [Google Scholar] [CrossRef]
- Hughes, S.R.; Kay, P.; Brown, L.E. Global synthesis and critical evaluation of pharmaceutical data sets collected from river systems. Environ. Sci. Technol. 2013, 47, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Martínez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Forslund, K.; Sunagawa, S.; Kultima, J.R.; Mende, D.R.; Arumugam, M.; Typas, A.; Bork, P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013, 23, 1163–1169. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Yang, X.; Qin, J.; Lu, N.; Cheng, G.; Wu, N.; Pan, Y.; Li, J.; Zhu, L.; Wang, X.; et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013, 4, 2151. [Google Scholar] [CrossRef] [Green Version]
- Sommer, M.O.A.; Dantas, G.; Church, G.M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 2009, 325, 1128–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buelow, E.; Gonzalez, T.B.; Versluis, D.; Oostdijk, E.A.; Ogilvie, L.A.; van Mourik, M.S.; Oosterink, E.; van Passel, M.W.; Smidt, H.; D’Andrea, M.M.; et al. Effects of selective digestive decontamination (SDD) on the gut resistome. J. Antimicrob. Chemother. 2014, 69, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Salyers, A.A.; Gupta, A.; Wang, Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004, 12, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Guiney, D.G., Jr.; Davis, C.E. Identification of a conjugative R plasmid in Bacteroides ochraceus capable of transfer to Escherichia coli. Nature 1978, 274, 181–182. [Google Scholar] [CrossRef] [PubMed]
- Privitera, G.; Dublanchet, A.; Sebald, M. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J. Infect. Dis. 1979, 139, 97–101. [Google Scholar] [CrossRef]
- Lanza, V.F.; Baquero, F.; Martínez, J.L.; Ramos-Ruíz, R.; González-Zorn, B.; Andremont, A.; Sánchez-Valenzuela, A.; Ehrlich, S.D.; Kennedy, S.; Ruppé, E.; et al. In-depth resistome analysis by targeted metagenomics. Microbiome 2018, 6, 11. [Google Scholar] [CrossRef]
- Loman, N.J.; Constantinidou, C.; Christner, M.; Rohde, H.; Chan, J.Z.; Quick, J.; Weir, J.C.; Quince, C.; Smith, G.P.; Betley, J.R.; et al. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA 2013, 309, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Kumar S, S.B. An Overview of Mechanisms and Emergence of Antimicrobials Drug Resistance. Adv. Anim. Vet. Sci. 2013, 1, 7–14. [Google Scholar]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, E.; Mayer, M.J.; Cotter, P.D.; Narbad, A. Gut microbiota as a source of novel antimicrobials. Gut Microbes 2019, 10, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Kadouri, D.E.; To, K.; Shanks, R.M.; Doi, Y. Predatory bacteria: A potential ally against multidrug-resistant Gram-negative pathogens. PLoS ONE 2013, 8, e63397. [Google Scholar] [CrossRef] [Green Version]
- Shatzkes, K.; Connell, N.D.; Kadouri, D.E. Predatory bacteria: A new therapeutic approach for a post-antibiotic era. Future Microbiol. 2017, 12, 469–472. [Google Scholar] [CrossRef]
- Shatzkes, K.; Tang, C.; Singleton, E.; Shukla, S.; Zuena, M.; Gupta, S.; Dharani, S.; Rinaggio, J.; Connell, N.D.; Kadouri, D.E. Effect of predatory bacteria on the gut bacterial microbiota in rats. Sci. Rep. 2017, 7, 43483. [Google Scholar] [CrossRef]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.H.; Anggård, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef]
- Yosef, I.; Manor, M.; Kiro, R.; Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 7267–7272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shatzkes, K.; Singleton, E.; Tang, C.; Zuena, M.; Shukla, S.; Gupta, S.; Dharani, S.; Onyile, O.; Rinaggio, J.; Connell, N.D.; et al. Predatory Bacteria Attenuate Klebsiella pneumoniae Burden in Rat Lungs. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Findlay, J.S.; Flick-Smith, H.C.; Keyser, E.; Cooper, I.A.; Williamson, E.D.; Oyston, P.C.F. Predatory bacteria can protect SKH-1 mice from a lethal plague challenge. Sci. Rep. 2019, 9, 7225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marine, E.; Milner, D.S.; Lambert, C.; Sockett, R.E.; Pos, K.M. A novel method to determine antibiotic sensitivity in Bdellovibrio bacteriovorus reveals a DHFR-dependent natural trimethoprim resistance. Sci. Rep. 2020, 10, 5315. [Google Scholar] [CrossRef] [Green Version]
- Madhusoodanan, J. Inner Workings: Probing predatory bacteria as an antibacterial remedy. Proc. Natl. Acad. Sci. USA 2019, 116, 22887–22890. [Google Scholar] [CrossRef] [Green Version]
- Cattoir, V.; Felden, B. Future Antibacterial Strategies: From Basic Concepts to Clinical Challenges. J. Infect Dis. 2019, 220, 350–360. [Google Scholar] [CrossRef]
- Chanishvili, N. Phage therapy—History from Twort and d’Herelle through Soviet experience to current approaches. Adv. Virus Res. 2012, 83, 3–40. [Google Scholar] [CrossRef]
- Watanabe, R.; Matsumoto, T.; Sano, G.; Ishii, Y.; Tateda, K.; Sumiyama, Y.; Uchiyama, J.; Sakurai, S.; Matsuzaki, S.; Imai, S.; et al. Efficacy of bacteriophage therapy against gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob. Agents Chemother. 2007, 51, 446–452. [Google Scholar] [CrossRef] [Green Version]
- Biswas, B.; Adhya, S.; Washart, P.; Paul, B.; Trostel, A.N.; Powell, B.; Carlton, R.; Merril, C.R. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect. Immun. 2002, 70, 204–210. [Google Scholar] [CrossRef] [Green Version]
- Ramesh, V.; Fralick, J.A.; Rolfe, R.D. Prevention of Clostridium difficile-induced ileocecitis with Bacteriophage. Anaerobe 1999, 5, 69–78. [Google Scholar] [CrossRef]
- Wang, J.; Hu, B.; Xu, M.; Yan, Q.; Liu, S.; Zhu, X.; Sun, Z.; Tao, D.; Ding, L.; Reed, E.; et al. Therapeutic effectiveness of bacteriophages in the rescue of mice with extended spectrum beta-lactamase-producing Escherichia coli bacteremia. Int. J. Mol. Med. 2006, 17, 347–355. [Google Scholar] [PubMed]
- Wills, Q.F.; Kerrigan, C.; Soothill, J.S. Experimental bacteriophage protection against Staphylococcus aureus abscesses in a rabbit model. Antimicrob. Agents Chemother. 2005, 49, 1220–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutateladze, M.; Adamia, R. Phage therapy experience at the Eliava Institute. Med. Mal. Infect. 2008, 38, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Fish, R.; Kutter, E.; Wheat, G.; Blasdel, B.; Kutateladze, M.; Kuhl, S. Bacteriophage treatment of intransigent diabetic toe ulcers: A case series. J. Wound Care 2016, 25. [Google Scholar] [CrossRef]
- Pouillot, F.; Chomton, M.; Blois, H.; Courroux, C.; Noelig, J.; Bidet, P.; Bingen, E.; Bonacorsi, S. Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b:H4-ST131 Escherichia coli strain producing CTX-M-15. Antimicrob. Agents Chemother. 2012, 56, 3568–3575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opal, S.M. Non-antibiotic treatments for bacterial diseases in an era of progressive antibiotic resistance. Crit. Care 2016, 20, 397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, S.M. Cytokine therapy of mycobacterial infections. Adv. Intern. Med. 2000, 45, 431–452. [Google Scholar]
- Bonanno, G.; Procoli, A.; Mariotti, A.; Corallo, M.; Perillo, A.; Danese, S.; De Cristofaro, R.; Scambia, G.; Rutella, S. Effects of pegylated G-CSF on immune cell number and function in patients with gynecological malignancies. J. Transl. Med. 2010, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Molineux, G. The design and development of pegfilgrastim (PEG-rmetHuG-CSF, Neulasta). Curr. Pharm. Des. 2004, 10, 1235–1244. [Google Scholar] [CrossRef]
- Canning, P.; Hassfurther, R.; TerHune, T.; Rogers, K.; Abbott, S.; Kolb, D. Efficacy and clinical safety of pegbovigrastim for preventing naturally occurring clinical mastitis in periparturient primiparous and multiparous cows on US commercial dairies. J. Dairy Sci. 2017, 100, 6504–6515. [Google Scholar] [CrossRef]
- Trimboli, F.; Morittu, V.M.; Di Loria, A.; Minuti, A.; Spina, A.A.; Piccioli-Cappelli, F.; Trevisi, E.; Britti, D.; Lopreiato, V. Effect of Pegbovigrastim on Hematological Profile of Simmental Dairy Cows during the Transition Period. Animals 2019, 9, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soliman, C.; Walduck, A.K.; Yuriev, E.; Richards, J.S.; Cywes-Bentley, C.; Pier, G.B.; Ramsland, P.A. Structural basis for antibody targeting of the broadly expressed microbial polysaccharide poly-N-acetylglucosamine. J. Biol. Chem. 2018, 293, 5079–5089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domenech, M.; Sempere, J.; de Miguel, S.; Yuste, J. Combination of Antibodies and Antibiotics as a Promising Strategy Against Multidrug-Resistant Pathogens of the Respiratory Tract. Front. Immunol. 2018, 9, 2700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navalkele, B.D.; Chopra, T. Bezlotoxumab: An emerging monoclonal antibody therapy for prevention of recurrent Clostridium difficile infection. Biologics 2018, 12, 11–21. [Google Scholar] [CrossRef] [Green Version]
- McCrea, K.; Ward, R.; LaRosa, S.P. Removal of Carbapenem-Resistant Enterobacteriaceae (CRE) from blood by heparin-functional hemoperfusion media. PLoS ONE 2014, 9, e114242. [Google Scholar] [CrossRef]
- Kang, J.H.; Super, M.; Yung, C.W.; Cooper, R.M.; Domansky, K.; Graveline, A.R.; Mammoto, T.; Berthet, J.B.; Tobin, H.; Cartwright, M.J.; et al. An extracorporeal blood-cleansing device for sepsis therapy. Nat. Med. 2014, 20, 1211–1216. [Google Scholar] [CrossRef]
- Amara, N.; Krom, B.P.; Kaufmann, G.F.; Meijler, M.M. Macromolecular inhibition of quorum sensing: Enzymes, antibodies, and beyond. Chem. Rev. 2011, 111, 195–208. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.H.; Zhang, L.H. Quorum sensing and quorum-quenching enzymes. J. Microbiol. 2005, 43, 101–109. [Google Scholar]
- McDougald, D.; Rice, S.A.; Kjelleberg, S. Bacterial quorum sensing and interference by naturally occurring biomimics. Anal. Bioanal. Chem. 2007, 387, 445–453. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.K.; Vinothkumar, K.; Rajpara, N. Bacterial quorum sensing inhibitors: Attractive alternatives for control of infectious pathogens showing multiple drug resistance. Recent. Pat. Antiinfect Drug Discov. 2013, 8, 68–83. [Google Scholar] [CrossRef]
- Dong, Y.H.; Wang, L.Y.; Zhang, L.H. Quorum-quenching microbial infections: Mechanisms and implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Aleksić, I.; Šegan, S.; Andrić, F.; Zlatović, M.; Moric, I.; Opsenica, D.M.; Senerovic, L. Long-Chain 4-Aminoquinolines as Quorum Sensing Inhibitors in Serratia marcescens and Pseudomonas aeruginosa. ACS Chem. Biol. 2017, 12, 1425–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalia, V.C.; Patel, S.K.S.; Kang, Y.C.; Lee, J.K. Quorum sensing inhibitors as antipathogens: Biotechnological applications. Biotechnol. Adv. 2019, 37, 68–90. [Google Scholar] [CrossRef] [PubMed]
- Fetzner, S. Quorum quenching enzymes. J. Biotechnol. 2015, 201, 2–14. [Google Scholar] [CrossRef]
- Cech, N.B.; Horswill, A.R. Small-molecule quorum quenchers to prevent Staphylococcus aureus infection. Future Microbiol. 2013, 8, 1511–1514. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Gao, Y.; Chen, X.; Yu, Z.; Li, X. Quorum quenching enzymes and their application in degrading signal molecules to block quorum sensing-dependent infection. Int. J. Mol. Sci. 2013, 14, 17477–17500. [Google Scholar] [CrossRef]
- Goswami, J. Quorum Sensing by Super Bugs and their Resistance to Antibiotics, a Short Review. Glob. J. Pharm. Pharm. Sci. 2017, 3. [Google Scholar]
- Brackman, G.; Coenye, T. Inhibition of Quorum Sensing in Staphylococcus spp. Curr. Pharm. Des. 2015, 21, 2101–2108. [Google Scholar] [CrossRef]
- Kalia, V.C. Quorum sensing inhibitors: An overview. Biotechnol. Adv. 2013, 31, 224–245. [Google Scholar] [CrossRef] [PubMed]
- Ejim, L.; Farha, M.A.; Falconer, S.B.; Wildenhain, J.; Coombes, B.K.; Tyers, M.; Brown, E.D.; Wright, G.D. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011, 7, 348–350. [Google Scholar] [CrossRef] [PubMed]
- Kalan, L.; Wright, G.D. Antibiotic adjuvants: Multicomponent anti-infective strategies. Expert Rev. Mol. Med. 2011, 13, e5. [Google Scholar] [CrossRef] [PubMed]
- Cottarel, G.; Wierzbowski, J. Combination drugs, an emerging option for antibacterial therapy. Trends Biotechnol. 2007, 25, 547–555. [Google Scholar] [CrossRef]
- Conrad, R.S.; Galanos, C. Fatty acid alterations and polymyxin B binding by lipopolysaccharides from Pseudomonas aeruginosa adapted to polymyxin B resistance. Antimicrob. Agents Chemother. 1989, 33, 1724–1728. [Google Scholar] [CrossRef] [Green Version]
- Speer, B.S.; Shoemaker, N.B.; Salyers, A.A. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 1992, 5, 387–399. [Google Scholar] [CrossRef]
- Van Bambeke, F.; Pagès, J.M.; Lee, V.J. Inhibitors of bacterial efflux pumps as adjuvants in antibiotic treatments and diagnostic tools for detection of resistance by efflux. Recent. Pat. Antiinfect. Drug Discov. 2006, 1, 157–175. [Google Scholar] [CrossRef] [Green Version]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. D-amino acids trigger biofilm disassembly. Science 2010, 328, 627–629. [Google Scholar] [CrossRef] [Green Version]
- Pérez, J.M.; Calderón, I.L.; Arenas, F.A.; Fuentes, D.E.; Pradenas, G.A.; Fuentes, E.L.; Sandoval, J.M.; Castro, M.E.; Elías, A.O.; Vásquez, C.C. Bacterial toxicity of potassium tellurite: Unveiling an ancient enigma. PLoS ONE 2007, 2, e211. [Google Scholar] [CrossRef]
- Calderón, I.L.; Elías, A.O.; Fuentes, E.L.; Pradenas, G.A.; Castro, M.E.; Arenas, F.A.; Pérez, J.M.; Vásquez, C.C. Tellurite-mediated disabling of [4Fe-4S] clusters of Escherichia coli dehydratases. Microbiology 2009, 155, 1840–1846. [Google Scholar] [CrossRef] [Green Version]
- Castro, M.E.; Molina, R.C.; Díaz, W.A.; Pradenas, G.A.; Vásquez, C.C. Expression of Aeromonas caviae ST pyruvate dehydrogenase complex components mediate tellurite resistance in Escherichia coli. Biochem. Biophys. Res. Commun. 2009, 380, 148–152. [Google Scholar] [CrossRef]
- Turner, R.J.; Aharonowitz, Y.; Weiner, J.H.; Taylor, D.E. Glutathione is a target in tellurite toxicity and is protected by tellurite resistance determinants in Escherichia coli. Can. J. Microbiol. 2001, 47, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Pérez, J.M.; Arenas, F.A.; Pradenas, G.A.; Sandoval, J.M.; Vásquez, C.C. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 2008, 283, 7346–7353. [Google Scholar] [CrossRef] [Green Version]
- Annunziato, G. Strategies to Overcome Antimicrobial Resistance (AMR) Making Use of Non-Essential Target Inhibitors: A Review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef] [Green Version]
- Douafer, H.; Andrieu, V.; Phanstiel, O.t.; Brunel, J.M. Antibiotic Adjuvants: Make Antibiotics Great Again! J. Med. Chem. 2019, 62, 8665–8681. [Google Scholar] [CrossRef]
- Chan, W.Y.; Khazandi, M.; Hickey, E.E.; Page, S.W.; Trott, D.J.; Hill, P.B. In vitro antimicrobial activity of seven adjuvants against common pathogens associated with canine otitis externa. Vet. Dermatol. 2019, 30, 133. [Google Scholar] [CrossRef]
- Bakken, J.S.; Borody, T.; Brandt, L.J.; Brill, J.V.; Demarco, D.C.; Franzos, M.A.; Kelly, C.; Khoruts, A.; Louie, T.; Martinelli, L.P.; et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011, 9, 1044–1049. [Google Scholar] [CrossRef] [Green Version]
- Smits, L.P.; Bouter, K.E.; de Vos, W.M.; Borody, T.J.; Nieuwdorp, M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology 2013, 145, 946–953. [Google Scholar] [CrossRef]
- DePeters, E.J.; George, L.W. Rumen transfaunation. Immunol. Lett. 2014, 162, 69–76. [Google Scholar] [CrossRef]
- Eiseman, B.; Silen, W.; Bascom, G.S.; Kauvar, A.J. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 1958, 44, 854–859. [Google Scholar]
- Zhang, F.; Luo, W.; Shi, Y.; Fan, Z.; Ji, G. Should we standardize the 1700-year-old fecal microbiota transplantation? Am. J. Gastroenterol. 2012. [Google Scholar] [CrossRef]
- Wortelboer, K.; Nieuwdorp, M.; Herrema, H. Fecal microbiota transplantation beyond Clostridioides difficile infections. EBioMedicine 2019, 44, 716–729. [Google Scholar] [CrossRef] [Green Version]
- Khoruts, A.; Staley, C.; Sadowsky, M.J. Faecal microbiota transplantation for Clostridioides difficile: Mechanisms and pharmacology. Nat. Rev. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef]
- Gopinath, P.M.; Narchonai, G.; Dhanasekaran, D.; Ranjani, A.; Thajuddin, N. Mycosynthesis, characterization and antibacterial properties of AgNPs against multidrug resistant (MDR) bacterial pathogens of female infertility cases. Asian J. Pharm. Sci. 2015, 10, 138–145. [Google Scholar] [CrossRef] [Green Version]
- Ingle, A.P.; Duran, N.; Rai, M. Bioactivity, mechanism of action, and cytotoxicity of copper-based nanoparticles: A review. Appl. Microbiol. Biotechnol. 2014, 98, 1001–1009. [Google Scholar] [CrossRef]
- Li, X.; Robinson, S.M.; Gupta, A.; Saha, K.; Jiang, Z.; Moyano, D.F.; Sahar, A.; Riley, M.A.; Rotello, V.M. Functional gold nanoparticles as potent antimicrobial agents against multi-drug-resistant bacteria. ACS Nano 2014, 8, 10682–10686. [Google Scholar] [CrossRef]
- Ibrahem, K.H.; Salman, J.A.S.; Ali, F.A. Effect of titanium nanoparticles biosynthesis by Lactobacillus crispatus on urease, hemolysin & biofilm forming by some bacteria causing recurrent UTI in Iraqi women. Eur. Sci. J. 2014, 10. [Google Scholar]
- Vincent, M.G.; John, N.P.; Narayanan, P.M.; Vani, C.; Murugan, S. In vitro study on the efficacy of zinc oxide and titanium dioxide nanoparticles against metallo beta-lactamase and biofilm producing Pseudomonas aeruginosa. J. Appl. Pharm. Sci. 2014, 4, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Malka, E.; Perelshtein, I.; Lipovsky, A.; Shalom, Y.; Naparstek, L.; Perkas, N.; Patick, T.; Lubart, R.; Nitzan, Y.; Banin, E.; et al. Eradication of multi-drug resistant bacteria by a novel Zn-doped CuO nanocomposite. Small 2013, 9, 4069–4076. [Google Scholar] [CrossRef]
- Huang, Z.; Zheng, X.; Yan, D.; Yin, G.; Liao, X.; Kang, Y.; Yao, Y.; Huang, D.; Hao, B. Toxicological effect of ZnO nanoparticles based on bacteria. Langmuir 2008, 24, 4140–4144. [Google Scholar] [CrossRef]
- Ansari, M.A.; Khan, H.M.; Khan, A.A.; Cameotra, S.S.; Saquib, Q.; Musarrat, J. Interaction of Al(2)O(3) nanoparticles with Escherichia coli and their cell envelope biomolecules. J. Appl. Microbiol. 2014, 116, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Moure, J.S.; Evangelopoulos, M.; Colvill, K.; Van Eps, J.L.; Tasciotti, E. Nanoantibiotics: A new paradigm for the treatment of surgical infection. Nanomedicine (Lond.) 2017, 12, 1319–1334. [Google Scholar] [CrossRef] [PubMed]
- Muzammil, S.; Hayat, S.; Fakhar, E.A.M.; Aslam, B.; Siddique, M.H.; Nisar, M.A.; Saqalein, M.; Atif, M.; Sarwar, A.; Khurshid, A.; et al. Nanoantibiotics: Future nanotechnologies to combat antibiotic resistance. Front. Biosci. (Elite Ed.) 2018, 10, 352–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witte, M.B.; Barbul, A. Role of nitric oxide in wound repair. Am. J. Surg. 2002, 183, 406–412. [Google Scholar] [CrossRef]
- Friedman, A.; Friedman, J. New biomaterials for the sustained release of nitric oxide: Past, present and future. Expert Opin. Drug Deliv. 2009, 6, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Kafshgari, M.H.; Cavallaro, A.; Delalat, B.; Harding, F.J.; McInnes, S.J.; Mäkilä, E.; Salonen, J.; Vasilev, K.; Voelcker, N.H. Nitric oxide-releasing porous silicon nanoparticles. Nanoscale Res. Lett. 2014, 9, 333. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Lee, D.G. PMAP-23 triggers cell death by nitric oxide-induced redox imbalance in Escherichia coli. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.S.; Kinnunen, A.; Karlsson, M.; Önnberg, A.; Söderquist, B.; Persson, K. The antibacterial effect of nitric oxide against ESBL-producing uropathogenic E. coli is improved by combination with miconazole and polymyxin B nonapeptide. BMC Microbiol. 2014, 14, 65. [Google Scholar] [CrossRef] [Green Version]
- Brisbois, E.J.; Bayliss, J.; Wu, J.; Major, T.C.; Xi, C.; Wang, S.C.; Bartlett, R.H.; Handa, H.; Meyerhoff, M.E. Optimized polymeric film-based nitric oxide delivery inhibits bacterial growth in a mouse burn wound model. Acta Biomater. 2014, 10, 4136–4142. [Google Scholar] [CrossRef] [Green Version]
- Kadam, S.; Shai, S.; Shahane, A.; Kaushik, K.S. Recent Advances in Non-Conventional Antimicrobial Approaches for Chronic Wound Biofilms: Have We Found the ’Chink in the Armor’? Biomedicines 2019, 7, 35. [Google Scholar] [CrossRef] [Green Version]
- Panacek, A.; Kvítek, L.; Prucek, R.; Kolar, M.; Vecerova, R.; Pizúrova, N.; Sharma, V.K.; Nevecna, T.; Zboril, R. Silver colloid nanoparticles: Synthesis, characterization, and their antibacterial activity. J. Phys. Chem. B 2006, 110, 16248–16253. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, M.M.; Elella, M.H.A.; Mohamed, R.R. Green synthesis of quaternized chitosan/silver nanocomposites for targeting mycobacterium tuberculosis and lung carcinoma cells (A-549). Int. J. Biol. Macromol. 2020, 142, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Saeb, A.T.; Alshammari, A.S.; Al-Brahim, H.; Al-Rubeaan, K.A. Production of silver nanoparticles with strong and stable antimicrobial activity against highly pathogenic and multidrug resistant bacteria. Sci. World J. 2014, 2014, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Panghal, M.; Kadyan, S.; Chaudhary, U.; Yadav, J.P. Green silver nanoparticles of Phyllanthus amarus: As an antibacterial agent against multi drug resistant clinical isolates of Pseudomonas aeruginosa. J. Nanobiotechnol. 2014, 12, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzoor-ul-Haq, V.R.; Singh, D.; Singh, A.K.; Ninganagouda, S.; Hiremath, A.J. Dried Mushroom Agaricus bisporus mediated synthesis of silver nanoparticles from Bandipora District (Jammu and Kashmir) and their efficacy against Methicillin Resistant Staphylococcus aureus (MRSA) strains. Nanosci. Nanotechnol. Int. J. 2015, 5, 1–8. [Google Scholar]
- Behera, S.; Nayak, P.L. In vitro Antibacterial Activity of Green Synthesized Silver Nanoparticles Using Jamun Extract Against Multiple Drug Resistant Bacteria. World J. Nano Sci. Technol. 2013, 2, 62–65. [Google Scholar] [CrossRef]
- Singh, K.; Panghal, M.; Kadyan, S.; Chaudhary, U.; Yadav, J.P. Antibacterial Activity of Synthesized Silver Nanoparticles from Tinospora cordifolia against Multi Drug Resistant Strains of Pseudomonas aeruginosa Isolated from Burn Patients. J. Nanomed. Nanotechnol. 2014, 5. [Google Scholar] [CrossRef]
- Chandrakanth, R.K.; Ashajyothi, C.; Oli, A.K.; Prabhurajeshwar, C. Potential Bactericidal Effect of Silver Nanoparticles Synthesised from Enterococcus Species. Orient. J. Chem. 2014, 30, 1253–1262. [Google Scholar]
- Lara, H.H.; Ayala-Nunez, N.V.; Ixtepan-Turrent, L.; Rodríguez-Padilla, C. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J. Microbiol. Biotechnol. 2010, 26, 615–621. [Google Scholar] [CrossRef]
- Ingle, A.; Gade, A.; Pierrat, S.; Sonnichsen, C.; Rai, M. Mycosynthesis of Silver Nanoparticles Using the Fungus Fusarium acuminatum and its Activity Against Some Human Pathogenic Bacteria. Curr. Nanosci. 2008, 4, 141–144. [Google Scholar] [CrossRef]
- Prabakar, K.; Sivalingam, P.; Mohamed Rabeek, S.I.; Muthuselvam, M.; Devarajan, N.; Arjunan, A.; Karthick, R.; Suresh, M.M.; Wembonyama, J.P. Evaluation of antibacterial efficacy of phyto fabricated silver nanoparticles using Mukia scabrella (Musumusukkai) against drug resistance nosocomial gram negative bacterial pathogens. Colloids Surf. B Biointerfaces 2013, 104, 282–288. [Google Scholar] [CrossRef] [PubMed]
- John, S.N. Evaluation of antibacteria lefficacy of phyto-fabricated gold nanoparticles using Bacopa monnieri plant extract. Indian J. Adv. Chem. Sci. 2013, 1, 94–98. [Google Scholar]
- Kunkalekar, R.K.; Naik, M.M.; Dubey, S.K.; Salker, A.V. Antibacterial activity of silver-dopedmanganese dioxide nanoparticles onmultidrug-resistant bacteria. J. Chem. Technol. Biotechnol. 2013, 88, 873–877. [Google Scholar] [CrossRef]
- Kalantar, E.; Kabir, K.; Gharibi, F.; Hatami, S.; Maleki, A. Effect and Properties of Surface-Modified Copper Doped ZnO Nanoparticles (Cu:ZnO NPs) on Killing Curves of Bacterial Pathogens. J. Med. Bacteriol. 2013, 2, 20–26. [Google Scholar]
- Bhande, R.M.; Khobragade, C.N.; Mane, R.S.; Bhande, S. Enhanced synergism of antibiotics with zinc oxide nanoparticles against extended spectrum b-lactamase producers implicated in urinary tract infections. J. Nanopart Res. 2013, 15, 1–13. [Google Scholar] [CrossRef]
- Roy, A.S.; Parveen, A.; Koppalkar, A.R.; Prasad, M.V.N.A. Effect of Nano - Titanium Dioxide with Different Antibiotics against Methicillin-Resistant Staphylococcus Aureus. J. Biomater. Nanobiotechnol. 2010, 1, 37–41. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.N.; Smith, K.; Samuels, T.A.; Lu, J.; Obare, S.O.; Scott, M.E. Nanoparticles functionalized with ampicillin destroy multiple-antibiotic-resistant isolates of Pseudomonas aeruginosa and Enterobacter aerogenes and methicillin-resistant Staphylococcus aureus. Appl. Environ. Microbiol. 2012, 78, 2768–2774. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, S.Z.; Kiran, U.; Ali, M.I.; Jamal, A.; Hameed, A.; Ahmed, S.; Ali, N. Combined efficacy of biologically synthesized silver nanoparticles and different antibiotics against multidrug-resistant bacteria. Int. J. Nanomed. 2013, 8, 3187–3195. [Google Scholar] [CrossRef] [Green Version]
- Burt, S. Essential oils: Their antibacterial properties and potential applications in foods--a review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [CrossRef] [Green Version]
- Savoia, D. Plant-derived antimicrobial compounds: Alternatives to antibiotics. Future Microbiol. 2012, 7, 979–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veras, H.N.; Rodrigues, F.F.; Colares, A.V.; Menezes, I.R.; Coutinho, H.D.; Botelho, M.A.; Costa, J.G. Synergistic antibiotic activity of volatile compounds from the essential oil of Lippia sidoides and thymol. Fitoterapia 2012, 83, 508–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raut, J.S.; Karuppayil, S.M. A status review on the medicinal properties of essential oils. Ind. Crops Prod. 2014, 62, 250–264. [Google Scholar] [CrossRef]
- Aleksic Sabo, V.; Knezevic, P. Antimicrobial activity of Eucalyptus camaldulensis Dehn. plant extracts and essential oils: A review. Ind. Crops Prod. 2019, 132, 413–429. [Google Scholar] [CrossRef]
- Wińska, K.; Mączka, W.; Łyczko, J.; Grabarczyk, M.; Czubaszek, A.; Szumny, A. Essential Oils as Antimicrobial Agents-Myth or Real Alternative? Molecules 2019, 24, 2130. [Google Scholar] [CrossRef] [Green Version]
- Mitscher, L.A.; Drake, S.; Gollapudi, S.R.; Okwute, S.K. A modern look at folkloric use of anti-infective agents. J. Nat. Prod. 1987, 50, 1025–1040. [Google Scholar] [CrossRef]
- Wang, Y.; Hong, J.; Liu, X.; Yang, H.; Liu, R.; Wu, J.; Wang, A.; Lin, D.; Lai, R. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PLoS ONE 2008, 3, e3217. [Google Scholar] [CrossRef] [Green Version]
- Flores-Villaseñor, H.; Canizalez-Román, A.; Reyes-Lopez, M.; Nazmi, K.; de la Garza, M.; Zazueta-Beltrán, J.; León-Sicairos, N.; Bolscher, J.G. Bactericidal effect of bovine lactoferrin, LFcin, LFampin and LFchimera on antibiotic-resistant Staphylococcus aureus and Escherichia coli. Biometals 2010, 23, 569–578. [Google Scholar] [CrossRef]
- Daniel, R. The soil metagenome—A rich resource for the discovery of novel natural products. Curr. Opin. Biotechnol. 2004, 15, 199–204. [Google Scholar] [CrossRef]
- Mokoena, M.P. Lactic Acid Bacteria and Their Bacteriocins: Classification, Biosynthesis and Applications against Uropathogens: A Mini-Review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef] [PubMed]
- Tsilingiri, K.; Barbosa, T.; Penna, G.; Caprioli, F.; Sonzogni, A.; Viale, G.; Rescigno, M. Probiotic and postbiotic activity in health and disease: Comparison on a novel polarised ex-vivo organ culture model. Gut 2012, 61, 1007–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar-Toalá, J.E.; Garcia-Varela, R.; Garcia, H.S.; Mata-Haro, V.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An evolving term within the functional foods field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Uyeno, Y.; Shigemori, S.; Shimosato, T. Effect of Probiotics/Prebiotics on Cattle Health and Productivity. Microbes Environ. 2015, 30, 126–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegh, C.A.M.; Geerlings, S.Y.; Knol, J.; Roeselers, G.; Belzer, C. Postbiotics and Their Potential Applications in Early Life Nutrition and Beyond. Int. J. Mol. Sci. 2019, 20, 4673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, A.K.; Malik, A. Antimicrobial potential and chemical composition of Mentha piperita oil in liquid and vapour phase against food spoiling microorganisms. Food Control 2011, 22, 1707–1714. [Google Scholar] [CrossRef]
- Mahboubi, M.; Haghi, G. Antimicrobial activity and chemical composition of Mentha pulegium L. essential oil. J. Ethnopharmacol. 2008, 119, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Khan, A.; Samber, N.; Manzoor, N. Antimicrobial activity of Mentha piperita essential oil in combination with silver ions. Synergy 2014, 1, 92–98. [Google Scholar] [CrossRef]
- Hussain, A.I.; Anwar, F.; Hussain Sherazi, S.T.; Przybylski, R. Chemical composition, antioxidant and antimicrobial activities of basil (Ocimum basilicum) essential oils depends on seasonal variations. Food Chem. 2008, 108, 986–995. [Google Scholar] [CrossRef]
- Opalchenova, G.; Obreshkova, D. Comparative studies on the activity of basil--an essential oil from Ocimum basilicum L.--against multidrug resistant clinical isolates of the genera Staphylococcus, Enterococcus and Pseudomonas by using different test methods. J. Microbiol. Methods 2003, 54, 105–110. [Google Scholar] [CrossRef]
- Koga, T.; Hirota, N.; Takumi, K. Bactericidal activities of essential oils of basil and sage against a range of bacteria and the effect of these essential oils on Vibrio parahaemolyticus. Microbiol. Res. 1999, 154, 267–273. [Google Scholar] [CrossRef]
- Sarikurkcu, C.; Zengin, G.; Oskay, M.; Uysal, S.; Ceylan, R.; Aktumsek, A. Composition, antioxidant, antimicrobial and enzyme inhibition activities of two Origanum vulgare subspecies (subsp. vulgare and subsp. hirtum) essential oils. Ind. Crops Prod. 2015, 70, 178–184. [Google Scholar] [CrossRef]
- Gutierrez, J.; Barry-Ryan, C.; Bourke, P. The antimicrobial efficacy of plant essential oil combinations and interactions with food ingredients. Int. J. Food Microbiol. 2008, 124, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Wu, N.; Fu, Y.-J.; Wang, W.; Luo, M.; Zhao, C.-J.; Zu, Y.-G.; Liu, X.-L. Chemical composition and antimicrobial activity of the essential oil of Rosemary. Environ. Toxicol. Pharmacol. 2011, 32, 63–68. [Google Scholar] [CrossRef]
- Ojeda-Sana, A.M.; van Baren, C.M.; Elechosa, M.A.; Juárez, M.A.; Moreno, S. New insights into antibacterial and antioxidant activities of rosemary essential oils and their main components. Food Control 2013, 31, 189–195. [Google Scholar] [CrossRef]
- Lemos, M.F.; Lemos, M.F.; Pacheco, H.P.; Endringer, D.C.; Scherer, R. Seasonality modifies rosemary’s composition and biological activity. Ind. Crops Prod. 2015, 70, 41–47. [Google Scholar] [CrossRef]
- Barreto, H.M.; Silva Filho, E.C.; Lima, E.d.O.; Coutinho, H.D.M.; Morais-Braga, M.F.B.; Tavares, C.C.A.; Tintino, S.R.; Rego, J.V.; de Abreu, A.P.L.; Lustosa, M.d.C.G.; et al. Chemical composition and possible use as adjuvant of the antibiotic therapy of the essential oil of Rosmarinus officinalis L. Ind. Crops Prod. 2014, 59, 290–294. [Google Scholar] [CrossRef]
- Parmeciano Di Noto, G.; Molina, M.C.; Quiroga, C. Insights Into Non-coding RNAs as Novel Antimicrobial Drugs. Front Genet. 2019, 10, 57. [Google Scholar] [CrossRef]
- Lipsitch, M.; Siber, G.R. How Can Vaccines Contribute to Solving the Antimicrobial Resistance Problem? mBio 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.G.; Adithan, C.; Harish, B.N.; Sujatha, S.; Roy, G.; Malini, A. Antimicrobial resistance in India: A review. J. Nat. Sci. Biol. Med. 2013, 4, 286–291. [Google Scholar] [CrossRef] [Green Version]
- Metz, M.; Shlaes, D.M. Eight more ways to deal with antibiotic resistance. Antimicrob. Agents Chemother. 2014, 58, 4253–4256. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Cosials, A.; Schulz, E.C.; Lambertsen, L.; Smyshlyaev, G.; Rojas-Cordova, C.; Forslund, K.; Karaca, E.; Bebel, A.; Bork, P.; Barabas, O. Transposase-DNA Complex Structures Reveal Mechanisms for Conjugative Transposition of Antibiotic Resistance. Cell 2018, 173, 208–220.e220. [Google Scholar] [CrossRef] [Green Version]



| Urgent | Serious | Concerning |
|---|---|---|
| 1. A. baumannii, P. aeruginosa, carbapenem-resistant 2. Clostridium difficile (CDIFF) 3. N. gonorrhoeae-3rd generation cephalosporin-resistant, fluoroquinolone-resistant 4. Carbapenem- and 3rd generation cephalosporin resistant Enterobacteriaceae: K. pneumonia, E. coli, Enterobacter spp., Serratia spp., Proteus spp., and Providencia spp., Morganella spp. | 1. Streptococcus pneumonia, penicillin-non-susceptible 2. Haemophilus influenzae, ampicillin-resistant 3. Shigella spp., fluoroquinolone-resistant 4. Enterococcus spp., vancomycin resistant 5. Multidrug-resistant Acinetobacter 6. Drug resistant Campylobacter 7.Extended-spectrum β-lactamase producing Enterobacteriae (ESBLs) 8. Multidrug-resistant P. aeruginosa 9. Drug-resistant non-typhoidal Salmonella 10. Drug-resistant Salmonella serotype Typhi 11. Drug resistant M. tuberculosis 12. Methicillin-resistant S. aureus (MRSA) | 1. Group B Streptococcus (GBS), clindamycin resistant 2. Group A Streptococcus (GAS), erythromycin resistant 3. S. aureus, vancomycin resistant |
| Mechanism of Action | Name of Antibiotic Families |
|---|---|
| Inhibition of protein synthesis | Tetracyclines, aminoglycosides, streptogramins, ketolides, macrolides, lincosamides, daptomycin |
| Inhibition of DNA synthesis | Fluoroquinolones, daptomycin |
| Inhibition of RNA synthesis | Rifampin and other metronidazoles, daptomycin |
| Inhibition of cell wall synthesis | Penicillins, cephalosporins, carbapenems, monobactams, glycopeptides |
| Disrupt functions of bacterial outer membrane | Daptomycin, polymyxin B, colistin, and lipopetides |
| Competitive inhibition of folic acid synthesis | Sulfonamides, trimethoprim |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.B.; Arnipalli, S.R.; Ziouzenkova, O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics 2020, 9, 688. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9100688
Kumar SB, Arnipalli SR, Ziouzenkova O. Antibiotics in Food Chain: The Consequences for Antibiotic Resistance. Antibiotics. 2020; 9(10):688. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9100688
Chicago/Turabian StyleKumar, Shashi B., Shanvanth R. Arnipalli, and Ouliana Ziouzenkova. 2020. "Antibiotics in Food Chain: The Consequences for Antibiotic Resistance" Antibiotics 9, no. 10: 688. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9100688






