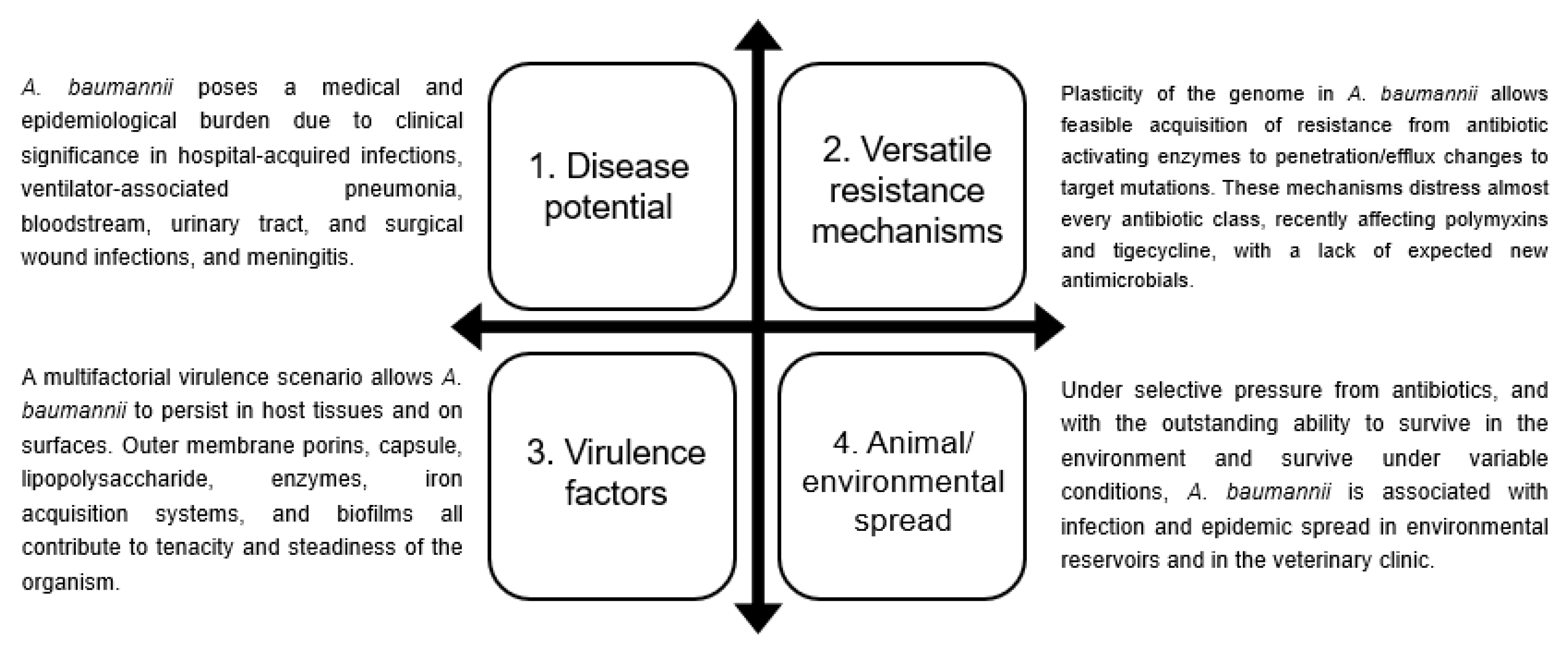
Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen
Abstract
:1. Introduction
2. Taxonomy and Microbiological Properties of Acinetobacter baumannii
3. Associated Infections and Clinical Impact of A. baumannii
3.1. Respiratory Infections
3.2. Bloodstream Infections
3.3. Skin and Soft Tissue Infections
3.4. Urinary Tract Infections
3.5. Meningitis
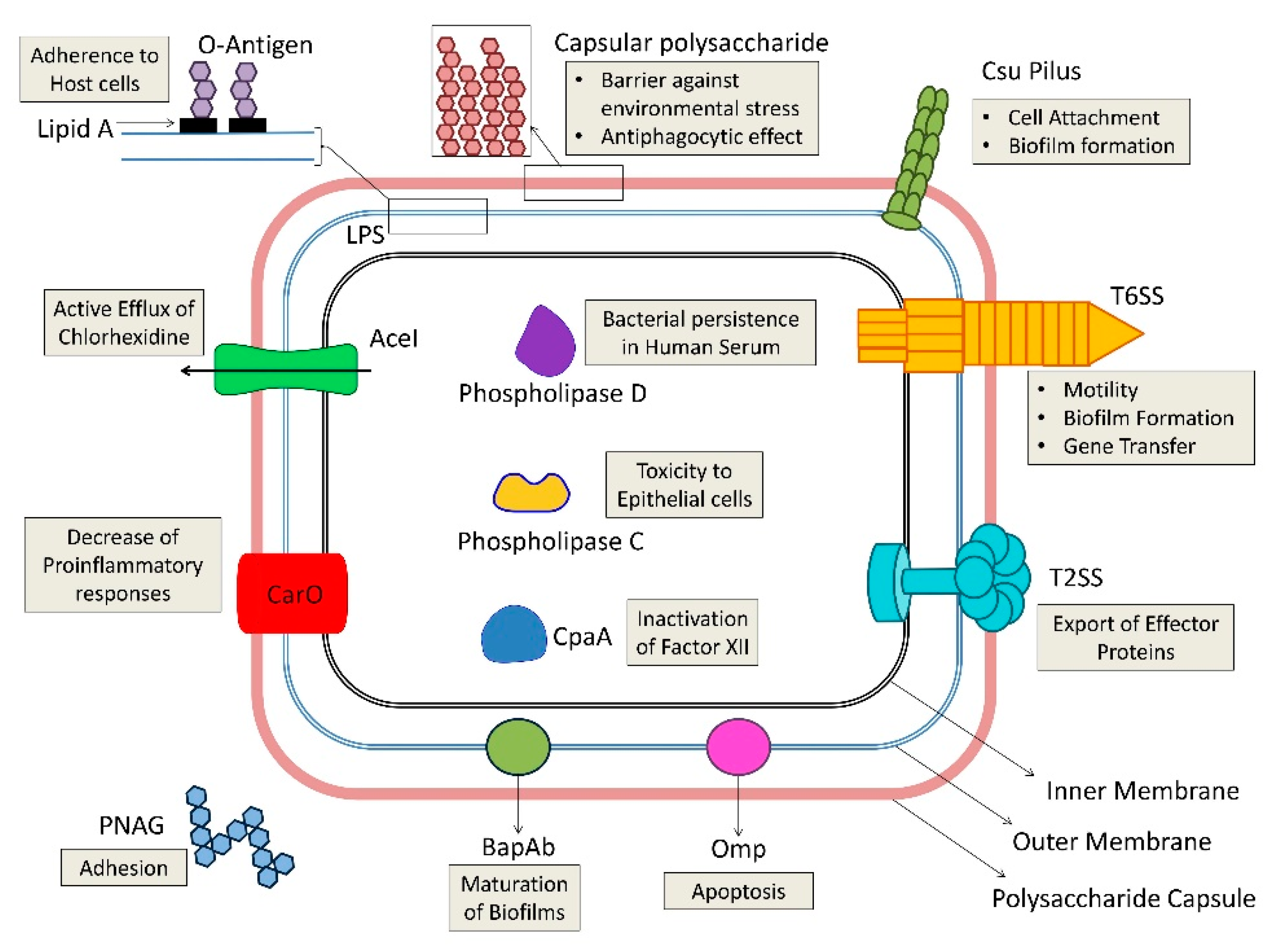
4. A. baumannii Virulence Factors
4.1. Outer Membrane Proteins (Porins)
4.2. Cell Envelope Factors (LPS and Capsule)
4.3. Enzymes
4.4. Capsular Polysaccharide Composition and Outer Membrane Resistance to Desiccation and Disinfection
4.5. Biofilm Production and Quorum Sensing
4.6. Motility
4.7. Micronutrient Acquisition Systems
4.8. Protein Secretion Systems
4.9. Others
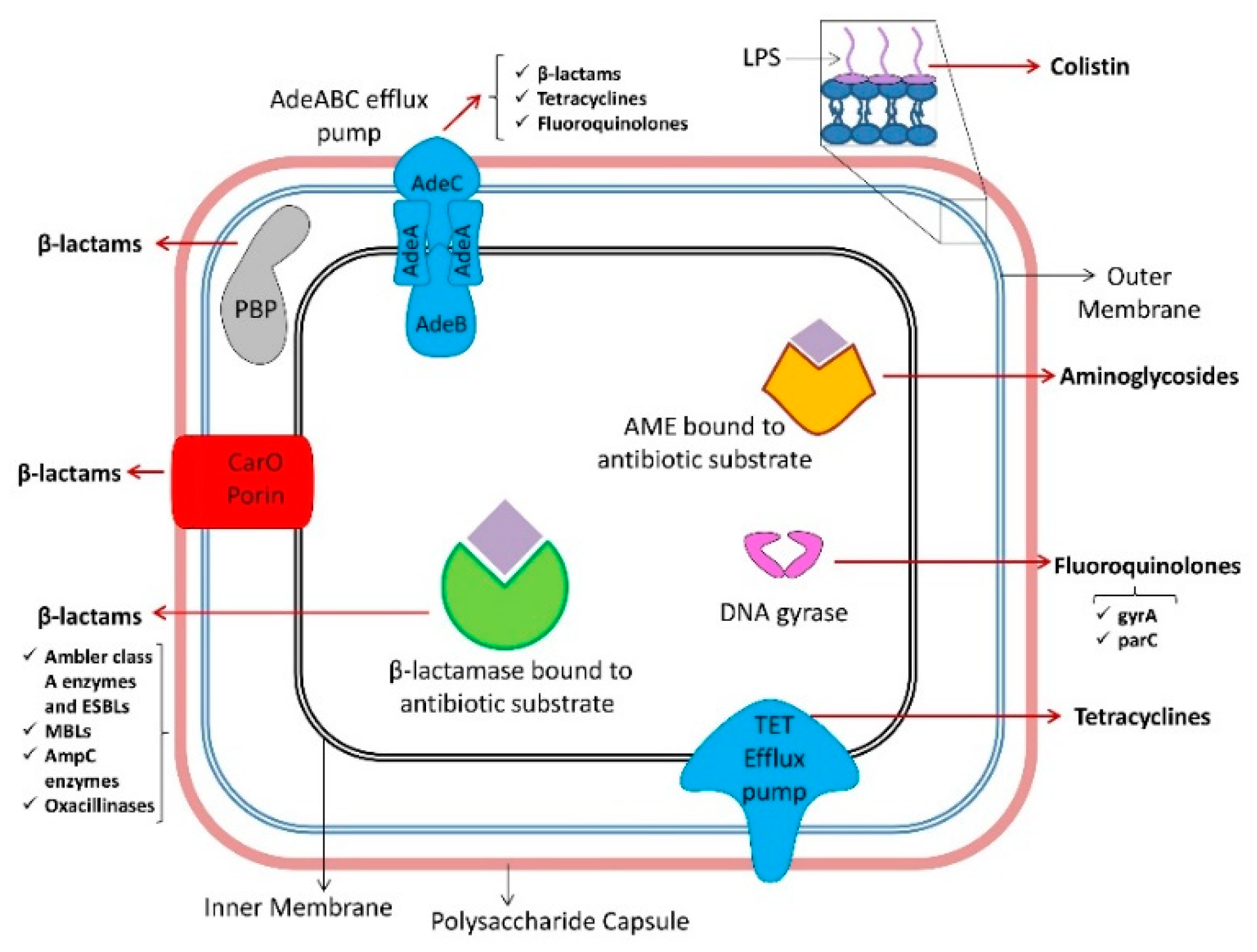
5. Mechanisms of Antibiotic Resistance in A. baumannii
5.1. β-lactams
5.1.1. Ambler Class A Enzymes
5.1.2. Ambler Class B Enzymes
5.1.3. Ambler Class C Enzymes
5.1.4. Ambler Class D Enzymes
5.1.5. Nonenzymatic β-lactam Resistance Mechanisms
5.2. Tetracyclines and Glycylcyclines
5.3. Fluoroquinolones
5.4. Aminoglycosides
5.5. Macrolides
5.6. Polymyxins
| Antibiotic | Resistance Mechanism | Enzyme/target/ Permeability Defect | Example | Reference |
|---|---|---|---|---|
| β-lactams | β-lactamases | Ambler class A | ESBLs of the families TEM, SHV, CTX-M, PER and VEB | [129,130,131,132] |
| SCO-1 (narrow spectrum) | [134] | |||
| Carbapenem-hydrolyzing ESBL of GES-type (GES-11) | [135,136] | |||
| KPC-2 | [139] | |||
| KPC-3 | [138,139] | |||
| KPC-5 | [137] | |||
| Ambler class B | NDM | [143,144,145,146,147] | ||
| VIM | [148] | |||
| GIM | [149] | |||
| SIM | [150] | |||
| IMP | [151,152,153] | |||
| Ambler class C | AmpC-69, AmpC-70, AmpC-71, and ADC-196 | [2,157,158] | ||
| Ambler class D | OXA-23-like | [20,136,164,165,167,170] | ||
| OXA24/40-like | [159] | |||
| OXA-51-like | [20,166] | |||
| OXA-58-like | [169] | |||
| OXA-143-like | [161] | |||
| OXA-235-like | [162] | |||
| Permeability lesions | Outer membrane porin downregulation | CarO | [171,172,174] | |
| OmpA, Omp33, OprB, Omp25, OprC, OprD, and OmpW | [175] | |||
| Efflux pump overactivity | RND pump | AdeABC | [2,176,177] | |
| Target mutation | PBP | PBP6b (dacD) | [173,174] | |
| Tertacyclines | Efflux pump overactivity | RND pump | AdeABC, AdeIJK, and AcrAB-TolC | [179,180,181,182,183,184,185] |
| Tet pump | TetA and TetB | [180] | ||
| Ribosomal protection | Tetracycline dissociation from ribosome | Tet(O) and Tet(M) | [180,186,187,188] | |
| Fluoroquinolones | Target mutation | DNA gyrase | GyrA | [191,192] |
| DNA topoisomerase IV | ParC | [191,192] | ||
| Efflux pump overactivity | RND pump | AdeABC | [81,191] | |
| MATE family | AbeM | [194] | ||
| Aminoglycosides | Drug inactivating enzymes | Aminoglycoside modifying enzymes | AAC(3)-Ia | [185] |
| AAC(3′)-Ia | [200] | |||
| ANT(2′)-Ia | [200] | |||
| ANT(3″)-Ia | [199] | |||
| Target mutation | 16sRNA methylase genes | armA, rmtA, rmtB, rmtC, and rmtD | [197,201] | |
| Efflux pump overactivity | RND pumps | AdeABC | [175,202] | |
| Macrolides | Efflux pump overactivity | SMR pump | AbeS | [204] |
| Polymyxins | Target mutation | Lipid A modification by PetN transferase | PmrC | [208,209,210] |
| MCR-1 | [214] | |||
| MCR-4 | [215,216] | |||
| Lack of lipid A biosynthesis | LpxA, LpxC, or LpxD | [211] | ||
| Decreased stability of outer membrane | LpsB, LptD, and VacJ | [212] | ||
| Reduced biotin synthesis | LpsB | [207,212,213] |
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Clark, N.M.; Zhanel, G.G.; Lynch, J.P. Emergence of antimicrobial resistance among Acinetobacter species: A global threat. Curr. Opin. Crit. Care 2016, 22, 491–499. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianco, A.; Quirino, A.; Giordano, M.; Marano, V.; Rizzo, C.; Liberto, M.C.; Focà, A.; Pavia, M. Control of carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit of a teaching hospital in Southern Italy. BMC Infect. Dis. 2016, 16, 747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Smiline Girija, A.S.; Priyadharsini, J.V. CLSI based antibiogram profile and the detection of MDR and XDR strains of Acinetobacter baumannii isolated from urine samples. Med. J. Islam. Repub. Iran 2019, 33, 3. [Google Scholar]
- Assimakopoulos, S.F.; Karamouzos, V.; Lefkaditi, A.; Sklavou, C.; Kolonitsiou, F.; Christofidou, M.; Fligou, F.; Gogos, C.; Marangos, M. Triple combination therapy with high-dose ampicillin/sulbactam, high-dose tigecycline and colistin in the treatment of ventilator-associated pneumonia caused by pan-drug resistant Acinetobacter baumannii: A case series study. Infez. Med. 2019, 27, 11–16. [Google Scholar]
- Goic-Barisic, I.; Seruga Music, M.; Kovacic, A.; Tonkic, M.; Hrenovic, J. Pan Drug-Resistant Environmental Isolate of Acinetobacter baumannii from Croatia. Microb. Drug Resist. 2017, 23, 494–496. [Google Scholar] [CrossRef] [Green Version]
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 21 August 2019).
- Bouvet, P.J.; Grimont, P.A. Identification and biotyping of clinical isolates of Acinetobacter. Ann. Inst. Pasteur Microbiol. 1987, 138, 569–578. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Biswas, I.; Veeraraghavan, B. Accurate identification of clinically important Acinetobacter spp.: An update. Future Sci. OA 2019, 5, FSO395. [Google Scholar] [CrossRef] [Green Version]
- Gerner-Smidt, P. Ribotyping of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Clin. Microbiol. 1992, 30, 2680–2685. [Google Scholar] [CrossRef] [Green Version]
- Nemec, A.; Krizova, L.; Maixnerova, M.; Sedo, O.; Brisse, S.; Higgins, P.G. Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2015, 65, 934–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosgaya, C.; Marí-Almirall, M.; Van Assche, A.; Fernández-Orth, D.; Mosqueda, N.; Telli, M.; Huys, G.; Higgins, P.G.; Seifert, H.; Lievens, B.; et al. Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int. J. Syst. Evol. Microbiol. 2016, 66, 4105–4111. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, X.; Zhao, W. Emerging Microtechnologies and Automated Systems for Rapid Bacterial Identification and Antibiotic Susceptibility Testing. SLAS Technol. 2017, 22, 585–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjernberg, I.; Ursing, J. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS Acta Pathol. Microbiol. Immunol. Scand. 1989, 97, 595–605. [Google Scholar] [CrossRef]
- Misbah, S.; Hassan, H.; Yusof, M.Y.; Hanifah, Y.A.; AbuBakar, S. Genomic species identification of Acinetobacter of clinical isolates by 16S rDNA sequencing. Singap. Med. J. 2005, 46, 461–464. [Google Scholar]
- Šedo, O.; Nemec, A.; Křížová, L.; Kačalová, M.; Zdráhal, Z. Improvement of MALDI-TOF MS profiling for the differentiation of species within the Acinetobacter calcoaceticus-Acinetobacter baumannii complex. Syst. Appl. Microbiol. 2013, 36, 572–578. [Google Scholar] [CrossRef]
- Rafei, R.; Osman, M.; Dabboussi, F.; Hamze, M. Update on the epidemiological typing methods for Acinetobacter baumannii. Future Microbiol. 2019, 14, 1065–1080. [Google Scholar] [CrossRef]
- Fitzpatrick, M.A.; Ozer, E.A.; Hauser, A.R. Utility of Whole-Genome Sequencing in Characterizing Acinetobacter Epidemiology and Analyzing Hospital Outbreaks. J. Clin. Microbiol. 2016, 54, 593–612. [Google Scholar] [CrossRef] [Green Version]
- Evans, B.A.; Amyes, S.G.B. OXA β-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef] [Green Version]
- Turton, J.F.; Woodford, N.; Glover, J.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006, 44, 2974–2976. [Google Scholar] [CrossRef] [Green Version]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djordjevic, Z.M.; Folic, M.M.; Folic, N.D.; Gajovic, N.; Gajovic, O.; Jankovic, S.M. Risk factors for hospital infections caused by carbapanem-resistant Acinetobacter baumannii. J. Infect. Dev. Ctries. 2016, 10, 1073–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baran, G.; Erbay, A.; Bodur, H.; Ongürü, P.; Akinci, E.; Balaban, N.; Cevik, M.A. Risk factors for nosocomial imipenem-resistant Acinetobacter baumannii infections. Int. J. Infect. Dis. 2008, 12, 16–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.-Y.; Hsu, S.-Y.; Hsu, J.-F.; Chen, C.-L.; Wang, Y.-H.; Chiu, C.-H. Risk factors and molecular epidemiology of Acinetobacter baumannii bacteremia in neonates. J. Microbiol. Immunol. Infect. 2018, 51, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Fukuta, Y.; Muder, R.R.; Agha, M.E.; Clarke, L.G.; Wagener, M.M.; Hensler, A.M.; Doi, Y. Risk factors for acquisition of multidrug-resistant Acinetobacter baumannii among cancer patients. Am. J. Infect. Control 2013, 41, 1249–1252. [Google Scholar] [CrossRef] [PubMed]
- Jaruratanasirikul, S.; Nitchot, W.; Wongpoowarak, W.; Samaeng, M.; Nawakitrangsan, M. Population pharmacokinetics and Monte Carlo simulations of sulbactam to optimize dosage regimens in patients with ventilator-associated pneumonia caused by Acinetobacter baumannii. Eur. J. Pharm. Sci. 2019, 136, 104940. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Zhanel, G.G.; Clark, N.M. Infections Due to Acinetobacter baumannii in the ICU: Treatment Options. Semin. Respir. Crit. Care Med. 2017, 38, 311–325. [Google Scholar]
- Huang, Y.; Zhou, Q.; Wang, W.; Huang, Q.; Liao, J.; Li, J.; Long, L.; Ju, T.; Zhang, Q.; Wang, H.; et al. Acinetobacter baumannii Ventilator-Associated Pneumonia: Clinical Efficacy of Combined Antimicrobial Therapy and in vitro Drug Sensitivity Test Results. Front. Pharmacol. 2019, 10, 92. [Google Scholar] [CrossRef]
- Kanafani, Z.A.; Zahreddine, N.; Tayyar, R.; Sfeir, J.; Araj, G.F.; Matar, G.M.; Kanj, S.S. Multi-drug resistant Acinetobacter species: A seven-year experience from a tertiary care center in Lebanon. Antimicrob. Resist. Infect. Control 2018, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Čiginskienė, A.; Dambrauskienė, A.; Rello, J.; Adukauskienė, D. Ventilator-Associated Pneumonia due to Drug-Resistant Acinetobacter baumannii: Risk Factors and Mortality Relation with Resistance Profiles, and Independent Predictors of In-Hospital Mortality. Medicina 2019, 55, 49. [Google Scholar] [CrossRef] [Green Version]
- Nowak, J.; Zander, E.; Stefanik, D.; Higgins, P.G.; Roca, I.; Vila, J.; McConnell, M.J.; Cisneros, J.M.; Seifert, H. MagicBullet Working Group WP4 High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 2017, 72, 3277–3282. [Google Scholar] [CrossRef] [PubMed]
- Mohd Sazlly Lim, S.; Zainal Abidin, A.; Liew, S.M.; Roberts, J.A.; Sime, F.B. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: A systematic review and meta-analysis. J. Infect. 2019, 79, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Hoang Quoc, C.; Nguyen Thi Phuong, T.; Nguyen Duc, H.; Tran Le, T.; Tran Thi Thu, H.; Nguyen Tuan, S.; Phan Trong, L. Carbapenemase Genes and Multidrug Resistance of Acinetobacter baumannii: A Cross Sectional Study of Patients with Pneumonia in Southern Vietnam. Antibiotics 2019, 8, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dexter, C.; Murray, G.L.; Paulsen, I.T.; Peleg, A.Y. Community-acquired Acinetobacter baumannii: Clinical characteristics, epidemiology and pathogenesis. Expert Rev. Anti-Infect. Ther. 2015, 13, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [PubMed] [Green Version]
- Jia, H.; Sun, Q.; Ruan, Z.; Xie, X. Characterization of a small plasmid carrying the carbapenem resistance gene blaOXA-72 from community-acquired Acinetobacter baumannii sequence type 880 in China. Infect. Drug Resist. 2019, 12, 1545–1553. [Google Scholar] [CrossRef] [Green Version]
- Wisplinghoff, H.; Paulus, T.; Lugenheim, M.; Stefanik, D.; Higgins, P.G.; Edmond, M.B.; Wenzel, R.P.; Seifert, H. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J. Infect. 2012, 64, 282–290. [Google Scholar] [CrossRef]
- Freire, M.P.; de Oliveira Garcia, D.; Garcia, C.P.; Campagnari Bueno, M.F.; Camargo, C.H.; Kono Magri, A.S.G.; Francisco, G.R.; Reghini, R.; Vieira, M.F.; Ibrahim, K.Y.; et al. Bloodstream infection caused by extensively drug-resistant Acinetobacter baumannii in cancer patients: High mortality associated with delayed treatment rather than with the degree of neutropenia. Clin. Microbiol. Infect. 2016, 22, 352–358. [Google Scholar] [CrossRef]
- Tsitsopoulos, P.P.; Iosifidis, E.; Antachopoulos, C.; Anestis, D.M.; Karantani, E.; Karyoti, A.; Papaevangelou, G.; Kyriazidis, E.; Roilides, E.; Tsonidis, C. Nosocomial bloodstream infections in neurosurgery: A 10-year analysis in a center with high antimicrobial drug-resistance prevalence. Acta Neurochir. (Wien) 2016, 158, 1647–1654. [Google Scholar] [CrossRef]
- Gong, Y.L.; Yang, Z.C.; Yin, S.P.; Liu, M.X.; Zhang, C.; Luo, X.Q.; Peng, Y.Z. Analysis of the pathogenic characteristics of 162 severely burned patients with bloodstream infection. Chin. J. Burns 2016, 32, 529–535. [Google Scholar]
- Motbainor, H.; Bereded, F.; Mulu, W. Multi-drug resistance of blood stream, urinary tract and surgical site nosocomial infections of Acinetobacter baumannii and Pseudomonas aeruginosa among patients hospitalized at Felegehiwot referral hospital, Northwest Ethiopia: A cross-sectional study. BMC Infect. Dis. 2020, 20, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papathanakos, G.; Andrianopoulos, I.; Papathanasiou, A.; Priavali, E.; Koulenti, D.; Koulouras, V. Colistin-Resistant Acinetobacter baumannii Bacteremia: A Serious Threat for Critically Ill Patients. Microorganisms 2020, 8, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, K.A.; Moran, K.A.; McAllister, C.K.; Gray, P.J. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg. Infect. Dis. 2005, 11, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.N.; Burns, T.C.; Hayda, R.A.; Hospenthal, D.R.; Murray, C.K. Infectious complications of open type III tibial fractures among combat casualties. Clin. Infect. Dis. 2007, 45, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Keen, E.F.; Murray, C.K.; Robinson, B.J.; Hospenthal, D.R.; Co, E.-M.A.; Aldous, W.K. Changes in the incidences of multidrug-resistant and extensively drug-resistant organisms isolated in a military medical center. Infect. Control Hosp. Epidemiol. 2010, 31, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, M.C.; Albrecht, M.A.; Griffith, M.E.; Murray, C.K.; Chung, K.K.; Horvath, E.E.; Ward, J.A.; Hospenthal, D.R.; Holcomb, J.B.; Wolf, S.E. Impact of Acinetobacter infection on the mortality of burn patients. J. Am. Coll. Surg. 2006, 203, 546–550. [Google Scholar] [CrossRef]
- Dallo, S.F.; Weitao, T. Insights into acinetobacter war-wound infections, biofilms, and control. Adv. Skin Wound Care 2010, 23, 169–174. [Google Scholar] [CrossRef]
- De Carvalho, V.C.; de Oliveira, P.R.D.; Dal-Paz, K.; de Paula, A.P.; Félix, C.D.S.; Lima, A.L.L.M. Gram-negative osteomyelitis: Clinical and microbiological profile. Braz. J. Infect. Dis. 2012, 16, 63–67. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Liu, Z.; Lin, Z.; Zhang, X.; Fu, W. Microbiologic characteristics of pathogenic bacteria from hospitalized trauma patients who survived Wenchuan earthquake. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 2529–2535. [Google Scholar] [CrossRef]
- Di Venanzio, G.; Flores-Mireles, A.L.; Calix, J.J.; Haurat, M.F.; Scott, N.E.; Palmer, L.D.; Potter, R.F.; Hibbing, M.E.; Friedman, L.; Wang, B.; et al. Urinary tract colonization is enhanced by a plasmid that regulates uropathogenic Acinetobacter baumannii chromosomal genes. Nat. Commun. 2019, 10, 2763. [Google Scholar] [CrossRef]
- Gaynes, R.; Edwards, J.R. National Nosocomial Infections Surveillance System Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 2005, 41, 848–854. [Google Scholar] [PubMed]
- Balfousias, T.; Apostolopoulos, A.; Angelis, S.; Filippou, D.; Maris, S. Pandrug-resistant Acinetobacter baumannii Infection Identified in a Non-intensive Care Unit Patient: A Case Study. Cureus 2019, 11, e6321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.K.; Lee, G.H.; Baek, J.Y.; Chung, D.R.; Peck, K.R.; Song, J.-H.; Ko, K.S. A single clone of Acinetobacter baumannii, ST22, is responsible for high antimicrobial resistance rates of Acinetobacter spp. isolates that cause bacteremia and urinary tract infections in Korea. Microb. Drug Resist. 2010, 16, 143–149. [Google Scholar] [CrossRef]
- Siegman-Igra, Y.; Bar-Yosef, S.; Gorea, A.; Avram, J. Nosocomial acinetobacter meningitis secondary to invasive procedures: Report of 25 cases and review. Clin. Infect. Dis. 1993, 17, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Goda, R.; Borkar, S.A.; Katiyar, V.; Agarwal, S.; Kumar, A.; Mohapatra, S.; Kapil, A.; Suri, A.; Kale, S.S. Outcome following postneurosurgical Acinetobacter meningitis: An institutional experience of 72 cases. Neurosurg. Focus 2019, 47, E8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Zhang, C.; Ye, S. Acinetobacter baumannii meningitis in children: A case series and literature review. Infection 2019, 47, 643–649. [Google Scholar] [CrossRef]
- Pan, S.; Huang, X.; Wang, Y.; Li, L.; Zhao, C.; Yao, Z.; Cui, W.; Zhang, G. Efficacy of intravenous plus intrathecal/intracerebral ventricle injection of polymyxin B for post-neurosurgical intracranial infections due to MDR/XDR Acinectobacter baumannii: A retrospective cohort study. Antimicrob. Resist. Infect. Control 2018, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.G.; Gianoulis, T.A.; Pukatzki, S.; Mekalanos, J.J.; Ornston, L.N.; Gerstein, M.; Snyder, M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21, 601–614. [Google Scholar] [CrossRef] [Green Version]
- Sahl, J.W.; Johnson, J.K.; Harris, A.D.; Phillippy, A.M.; Hsiao, W.W.; Thom, K.A.; Rasko, D.A. Genomic comparison of multi-drug resistant invasive and colonizing Acinetobacter baumannii isolated from diverse human body sites reveals genomic plasticity. BMC Genom. 2011, 12, 291. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Huang, S.; Zhang, Q. Outer membrane proteins: Key players for bacterial adaptation in host niches. Microbes Infect. 2002, 4, 325–331. [Google Scholar] [CrossRef]
- Choi, C.H.; Lee, E.Y.; Lee, Y.C.; Park, T.I.; Kim, H.J.; Hyun, S.H.; Kim, S.A.; Lee, S.-K.; Lee, J.C. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 2005, 7, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.H.; Lee, J.S.; Lee, Y.C.; Park, T.I.; Lee, J.C. Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 2008, 8, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smani, Y.; McConnell, M.J.; Pachón, J. Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PLoS ONE 2012, 7, e33073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smani, Y.; Dominguez-Herrera, J.; Pachón, J. Association of the outer membrane protein Omp33 with fitness and virulence of Acinetobacter baumannii. J. Infect. Dis. 2013, 208, 1561–1570. [Google Scholar] [CrossRef] [Green Version]
- Rumbo, C.; Tomás, M.; Fernández Moreira, E.; Soares, N.C.; Carvajal, M.; Santillana, E.; Beceiro, A.; Romero, A.; Bou, G. The Acinetobacter baumannii Omp33-36 porin is a virulence factor that induces apoptosis and modulates autophagy in human cells. Infect. Immun. 2014, 82, 4666–4680. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Unno, Y.; Kawakami, S.; Ubagai, T.; Ono, Y. Virulence characteristics of Acinetobacter baumannii clinical isolates vary with the expression levels of omps. J. Med. Microbiol. 2017, 66, 203–212. [Google Scholar] [CrossRef]
- Knapp, S.; Wieland, C.W.; Florquin, S.; Pantophlet, R.; Dijkshoorn, L.; Tshimbalanga, N.; Akira, S.; van der Poll, T. Differential roles of CD14 and toll-like receptors 4 and 2 in murine Acinetobacter pneumonia. Am. J. Respir. Crit. Care Med. 2006, 173, 122–129. [Google Scholar] [CrossRef]
- Haseley, S.R.; Pantophlet, R.; Brade, L.; Holst, O.; Brade, H. Structural and serological characterisation of the O-antigenic polysaccharide of the lipopolysaccharide from Acinetobacter junii strain 65. Eur. J. Biochem. 1997, 245, 477–481. [Google Scholar] [CrossRef]
- Singh, J.K.; Adams, F.G.; Brown, M.H. Diversity and Function of Capsular Polysaccharide in Acinetobacter baumannii. Front. Microbiol. 2018, 9, 3301. [Google Scholar] [CrossRef]
- Geisinger, E.; Isberg, R.R. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015, 11, e1004691. [Google Scholar] [CrossRef] [Green Version]
- Kenyon, J.J.; Hall, R.M. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS ONE 2013, 8, e62160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camarena, L.; Bruno, V.; Euskirchen, G.; Poggio, S.; Snyder, M. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 2010, 6, e1000834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waack, U.; Warnock, M.; Yee, A.; Huttinger, Z.; Smith, S.; Kumar, A.; Deroux, A.; Ginsburg, D.; Mobley, H.L.T.; Lawrence, D.A.; et al. CpaA Is a Glycan-Specific Adamalysin-like Protease Secreted by Acinetobacter baumannii That Inactivates Coagulation Factor XII. mBio 2018, 9, e01606-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannouli, M.; Antunes, L.C.S.; Marchetti, V.; Triassi, M.; Visca, P.; Zarrilli, R. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect. Dis. 2013, 13, 282. [Google Scholar] [CrossRef] [Green Version]
- Ophir, T.; Gutnick, D.L. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 1994, 60, 740–745. [Google Scholar] [CrossRef] [Green Version]
- Boll, J.M.; Tucker, A.T.; Klein, D.R.; Beltran, A.M.; Brodbelt, J.S.; Davies, B.W.; Trent, M.S. Reinforcing Lipid A Acylation on the Cell Surface of Acinetobacter baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival. mBio 2015, 6, e00478-415. [Google Scholar] [CrossRef] [Green Version]
- Hassan, K.A.; Jackson, S.M.; Penesyan, A.; Patching, S.G.; Tetu, S.G.; Eijkelkamp, B.A.; Brown, M.H.; Henderson, P.J.F.; Paulsen, I.T. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 20254–20259. [Google Scholar] [CrossRef] [Green Version]
- Nwugo, C.C.; Arivett, B.A.; Zimbler, D.L.; Gaddy, J.A.; Richards, A.M.; Actis, L.A. Effect of ethanol on differential protein production and expression of potential virulence functions in the opportunistic pathogen Acinetobacter baumannii. PLoS ONE 2012, 7, e51936. [Google Scholar] [CrossRef] [Green Version]
- Asplund, M.B.; Coelho, C.; Cordero, R.J.B.; Martinez, L.R. Alcohol impairs J774.16 macrophage-like cell antimicrobial functions in Acinetobacter baumannii infection. Virulence 2013, 4, 467–472. [Google Scholar] [CrossRef] [Green Version]
- Doi, Y.; Murray, G.L.; Peleg, A.Y. Acinetobacter baumannii: Evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 2015, 36, 85–98. [Google Scholar]
- Greene, C.; Wu, J.; Rickard, A.H.; Xi, C. Evaluation of the ability of Acinetobacter baumannii to form biofilms on six different biomedical relevant surfaces. Lett. Appl. Microbiol. 2016, 63, 233–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomaras, A.P.; Flagler, M.J.; Dorsey, C.W.; Gaddy, J.A.; Actis, L.A. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiol. Read. Engl. 2008, 154, 3398–3409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loehfelm, T.W.; Luke, N.R.; Campagnari, A.A. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 2008, 190, 1036–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, H.M.S.; Beatson, S.A.; Totsika, M.; Moriel, D.G.; Phan, M.-D.; Szubert, J.; Runnegar, N.; Sidjabat, H.E.; Paterson, D.L.; Nimmo, G.R.; et al. Molecular analysis of the Acinetobacter baumannii biofilm-associated protein. Appl. Environ. Microbiol. 2013, 79, 6535–6543. [Google Scholar] [CrossRef] [Green Version]
- Choi, A.H.K.; Slamti, L.; Avci, F.Y.; Pier, G.B.; Maira-Litrán, T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 2009, 191, 5953–5963. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Saipriya, K.; Swathi, C.H.; Ratnakar, K.S.; Sritharan, V. Quorum-sensing system in Acinetobacter baumannii: A potential target for new drug development. J. Appl. Microbiol. 2019, 128, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Alarcon, I.; Evans, D.J.; Fleiszig, S.M.J. The role of twitching motility in Pseudomonas aeruginosa exit from and translocation of corneal epithelial cells. Investig. Ophthalmol. Vis. Sci. 2009, 50, 2237–2244. [Google Scholar] [CrossRef] [Green Version]
- Skiebe, E.; de Berardinis, V.; Morczinek, P.; Kerrinnes, T.; Faber, F.; Lepka, D.; Hammer, B.; Zimmermann, O.; Ziesing, S.; Wichelhaus, T.A.; et al. Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int. J. Med. Microbiol. IJMM 2012, 302, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Vijayakumar, S.; Rajenderan, S.; Laishram, S.; Anandan, S.; Balaji, V.; Biswas, I. Biofilm Formation and Motility Depend on the Nature of the Acinetobacter baumannii Clinical Isolates. Front. Public Health 2016, 4, 105. [Google Scholar] [CrossRef]
- Harding, C.M.; Tracy, E.N.; Carruthers, M.D.; Rather, P.N.; Actis, L.A.; Munson, R.S. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 2013, 4, e00360-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilharm, G.; Piesker, J.; Laue, M.; Skiebe, E. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J. Bacteriol. 2013, 195, 4146–4153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrows, L.L. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu. Rev. Microbiol. 2012, 66, 493–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eijkelkamp, B.A.; Stroeher, U.H.; Hassan, K.A.; Elbourne, L.D.H.; Paulsen, I.T.; Brown, M.H. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect. Immun. 2013, 81, 2574–2583. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Varela, M.; Corral, J.; Vallejo, J.A.; Rumbo-Feal, S.; Bou, G.; Aranda, J.; Barbé, J. Mutations in the β-Subunit of the RNA Polymerase Impair the Surface-Associated Motility and Virulence of Acinetobacter baumannii. Infect. Immun. 2017, 85, e00327-17. [Google Scholar]
- Ahmad, I.; Nygren, E.; Khalid, F.; Myint, S.L.; Uhlin, B.E. A Cyclic-di-GMP signalling network regulates biofilm formation and surface associated motility of Acinetobacter baumannii 17978. Sci. Rep. 2020, 10, 1991. [Google Scholar] [CrossRef]
- Tipton, K.A.; Rather, P.N. An ompR-envZ Two-Component System Ortholog Regulates Phase Variation, Osmotic Tolerance, Motility, and Virulence in Acinetobacter baumannii Strain AB5075. J. Bacteriol. 2017, 199, e00705-16. [Google Scholar] [CrossRef] [Green Version]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The Mechanisms of Disease Caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef] [Green Version]
- Ajiboye, T.O.; Skiebe, E.; Wilharm, G. Contributions of ferric uptake regulator Fur to the sensitivity and oxidative response of Acinetobacter baumannii to antibiotics. Microb. Pathog. 2018, 119, 35–41. [Google Scholar] [CrossRef]
- Moore, J.L.; Becker, K.W.; Nicklay, J.J.; Boyd, K.L.; Skaar, E.P.; Caprioli, R.M. Imaging mass spectrometry for assessing temporal proteomics: Analysis of calprotectin in Acinetobacter baumannii pulmonary infection. Proteomics 2014, 14, 820–828. [Google Scholar] [CrossRef] [Green Version]
- Nairn, B.L.; Lonergan, Z.R.; Wang, J.; Braymer, J.J.; Zhang, Y.; Calcutt, M.W.; Lisher, J.P.; Gilston, B.A.; Chazin, W.J.; de Crécy-Lagard, V.; et al. The Response of Acinetobacter baumannii to Zinc Starvation. Cell Host Microbe 2016, 19, 826–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juttukonda, L.J.; Chazin, W.J.; Skaar, E.P. Acinetobacter baumannii Coordinates Urea Metabolism with Metal Import To Resist Host-Mediated Metal Limitation. mBio 2016, 7, e01475-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, B.S.; Kinsella, R.L.; Harding, C.M.; Feldman, M.F. The Secrets of Acinetobacter Secretion. Trends Microbiol. 2017, 25, 532–545. [Google Scholar] [CrossRef]
- Bentancor, L.V.; Camacho-Peiro, A.; Bozkurt-Guzel, C.; Pier, G.B.; Maira-Litrán, T. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J. Bacteriol. 2012, 194, 3950–3960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandkvist, M. Type II secretion and pathogenesis. Infect. Immun. 2001, 69, 3523–3535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilley, D.; Law, R.; Warren, S.; Samis, J.A.; Kumar, A. CpaA a novel protease from Acinetobacter baumannii clinical isolates deregulates blood coagulation. FEMS Microbiol. Lett. 2014, 356, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Hood, R.D.; Singh, P.; Hsu, F.; Güvener, T.; Carl, M.A.; Trinidad, R.R.S.; Silverman, J.M.; Ohlson, B.B.; Hicks, K.G.; Plemel, R.L.; et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 2010, 7, 25–37. [Google Scholar] [CrossRef] [Green Version]
- Elhosseiny, N.M.; Attia, A.S. Acinetobacter: An emerging pathogen with a versatile secretome. Emerg. Microbes Infect. 2018, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Repizo, G.D. Prevalence of Acinetobacter baumannii strains expressing the Type 6 secretion system in patients with bacteremia. Virulence 2017, 8, 1099–1101. [Google Scholar] [CrossRef] [Green Version]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Fishbain, J.; Peleg, A.Y. Treatment of Acinetobacter infections. Clin. Infect. Dis. 2010, 51, 79–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garnacho-Montero, J.; Ortiz-Leyba, C.; Jiménez-Jiménez, F.J.; Barrero-Almodóvar, A.E.; García-Garmendia, J.L.; Bernabeu-WittelI, M.; Gallego-Lara, S.L.; Madrazo-Osuna, J. Treatment of multidrug-resistant Acinetobacter baumannii ventilator-associated pneumonia (VAP) with intravenous colistin: A comparison with imipenem-susceptible VAP. Clin. Infect. Dis. 2003, 36, 1111–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Superti, S.V.; Martins, D.D.S.; Caierão, J.; Soares, F.D.S.; Prochnow, T.; Zavascki, A.P. Indications of carbapenem resistance evolution through heteroresistance as an intermediate stage in Acinetobacter baumannii after carbapenem administration. Rev. Inst. Med. Trop. Sao Paulo 2009, 51, 111–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falagas, M.E.; Kasiakou, S.K. Toxicity of polymyxins: A systematic review of the evidence from old and recent studies. Crit. Care Lond. Engl. 2006, 10, R27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, D.J.; Garavaglia-Wilson, A. A review of intravenous minocycline for treatment of multidrug-resistant Acinetobacter infections. Clin. Infect. Dis. 2014, 59 (Suppl. 6), S374–S380. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-T.; Tsao, S.-M.; Hsueh, P.-R. Clinical outcomes of tigecycline alone or in combination with other antimicrobial agents for the treatment of patients with healthcare-associated multidrug-resistant Acinetobacter baumannii infections. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1211–1220. [Google Scholar] [CrossRef]
- Karageorgopoulos, D.E.; Kelesidis, T.; Kelesidis, I.; Falagas, M.E. Tigecycline for the treatment of multidrug-resistant (including carbapenem-resistant) Acinetobacter infections: A review of the scientific evidence. J. Antimicrob. Chemother. 2008, 62, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Freire, A.T.; Melnyk, V.; Kim, M.J.; Datsenko, O.; Dzyublik, O.; Glumcher, F.; Chuang, Y.-C.; Maroko, R.T.; Dukart, G.; Cooper, C.A.; et al. Comparison of tigecycline with imipenem/cilastatin for the treatment of hospital-acquired pneumonia. Diagn. Microbiol. Infect. Dis. 2010, 68, 140–151. [Google Scholar] [CrossRef]
- Penwell, W.F.; Shapiro, A.B.; Giacobbe, R.A.; Gu, R.-F.; Gao, N.; Thresher, J.; McLaughlin, R.E.; Huband, M.D.; DeJonge, B.L.M.; Ehmann, D.E.; et al. Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2015, 59, 1680–1689. [Google Scholar] [CrossRef] [Green Version]
- Rigatto, M.H.; Vieira, F.J.; Antochevis, L.C.; Behle, T.F.; Lopes, N.T.; Zavascki, A.P. Polymyxin B in Combination with Antimicrobials Lacking In Vitro Activity versus Polymyxin B in Monotherapy in Critically Ill Patients with Acinetobacter baumannii or Pseudomonas aeruginosa Infections. Antimicrob. Agents Chemother. 2015, 59, 6575–6580. [Google Scholar] [CrossRef] [Green Version]
- Durante-Mangoni, E.; Signoriello, G.; Andini, R.; Mattei, A.; De Cristoforo, M.; Murino, P.; Bassetti, M.; Malacarne, P.; Petrosillo, N.; Galdieri, N.; et al. Colistin and rifampicin compared with colistin alone for the treatment of serious infections due to extensively drug-resistant Acinetobacter baumannii: A multicenter, randomized clinical trial. Clin. Infect. Dis. 2013, 57, 349–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spellberg, B.; Bonomo, R.A. Combination Therapy for Extreme Drug-Resistant Acinetobacter baumannii: Ready for Prime Time? Crit. Care Med. 2015, 43, 1332–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Minh, V.; Thi Khanh Nhu, N.; Vinh Phat, V.; Thompson, C.; Huong Lan, N.P.; Thieu Nga, T.V.; Thanh Tam, P.T.; Tuyen, H.T.; Hoang Nhu, T.D.; Van Hao, N.; et al. In vitro activity of colistin in antimicrobial combination against carbapenem-resistant Acinetobacter baumannii isolated from patients with ventilator-associated pneumonia in Vietnam. J. Med. Microbiol. 2015, 64, 1162–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oleksiuk, L.M.; Nguyen, M.H.; Press, E.G.; Updike, C.L.; O’Hara, J.A.; Doi, Y.; Clancy, C.J.; Shields, R.K. In vitro responses of Acinetobacter baumannii to two- and three-drug combinations following exposure to colistin and doripenem. Antimicrob. Agents Chemother. 2014, 58, 1195–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Lee, J.S.; Park, S.Y.; Ko, Y.; Eom, J.S. Colistin Plus Carbapenem versus Colistin Monotherapy in the Treatment of Carbapenem-Resistant Acinetobacter baumannii Pneumonia. Infect. Drug Resist. 2019, 12, 3925–3934. [Google Scholar] [CrossRef] [Green Version]
- Sertcelik, A.; Baran, I.; Akinci, E.; Mumcuoglu, I.; Bodur, H. Synergistic Activities of Colistin Combinations with Meropenem, Sulbactam, Minocycline, Disodium Fosfomycin, or Vancomycin Against Different Clones of Carbapenem-Resistant Acinetobacter baumannii Strains. Microb. Drug Resist. 2019. [Google Scholar] [CrossRef]
- Hammoudi, D.; Moubareck, C.A.; Sarkis, D.K. How to detect carbapenemase producers? A literature review of phenotypic and molecular methods. J. Microbiol. Methods 2014, 107, 106–118. [Google Scholar] [CrossRef]
- Alkasaby, N.M.; El Sayed Zaki, M. Molecular Study of Acinetobacter baumannii Isolates for Metallo-β-Lactamases and Extended-Spectrum-β-Lactamases Genes in Intensive Care Unit, Mansoura University Hospital, Egypt. Int. J. Microbiol. 2017, 2017, 3925868. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, Y.; Hunfeld, K.-P.; Borgmann, S.; Maneg, D.; Blobner, W.; Werner, G.; Higgins, P.G. Carbapenem-resistant Acinetobacter baumannii ST78 with OXA-72 carbapenemase and ESBL gene blaCTX-M-115. J. Antimicrob. Chemother. 2016, 71, 1426–1428. [Google Scholar] [CrossRef] [Green Version]
- Uddin, F.; McHugh, T.D.; Roulston, K.; Platt, G.; Khan, T.A.; Sohail, M. Detection of carbapenemases, AmpC and ESBL genes in Acinetobacter isolates from ICUs by DNA microarray. J. Microbiol. Methods 2018, 155, 19–23. [Google Scholar] [CrossRef]
- Safari, M.; Mozaffari Nejad, A.S.; Bahador, A.; Jafari, R.; Alikhani, M.Y. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU). Saudi J. Biol. Sci. 2015, 22, 424–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.-R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.-J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirel, L.; Corvec, S.; Rapoport, M.; Mugnier, P.; Petroni, A.; Pasteran, F.; Faccone, D.; Galas, M.; Drugeon, H.; Cattoir, V.; et al. Identification of the novel narrow-spectrum beta-lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob. Agents Chemother. 2007, 51, 2179–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moubareck, C.; Brémont, S.; Conroy, M.-C.; Courvalin, P.; Lambert, T. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 3579–3581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammoudi, D.; Moubareck, C.A.; Hakime, N.; Houmani, M.; Barakat, A.; Najjar, Z.; Suleiman, M.; Fayad, N.; Sarraf, R.; Sarkis, D.K. Spread of imipenem-resistant Acinetobacter baumannii co-expressing OXA-23 and GES-11 carbapenemases in Lebanon. Int. J. Infect. Dis. 2015, 36, 56–61. [Google Scholar] [CrossRef] [Green Version]
- Robledo, I.E.; Aquino, E.E.; Santé, M.I.; Santana, J.L.; Otero, D.M.; León, C.F.; Vázquez, G.J. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob. Agents Chemother. 2010, 54, 1354–1357. [Google Scholar] [CrossRef] [Green Version]
- Caneiras, C.; Calisto, F.; Jorge da Silva, G.; Lito, L.; Melo-Cristino, J.; Duarte, A. First Description of Colistin and Tigecycline-Resistant Acinetobacter baumannii Producing KPC-3 Carbapenemase in Portugal. Antibiotics 2018, 7, 96. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, P.C.S.; Monteiro, A.S.; Marques, S.G.; Monteiro, S.G.; Monteiro-Neto, V.; Coqueiro, M.M.M.; Marques, A.C.G.; de Jesus Gomes Turri, R.; Santos, S.G.; Bomfim, M.R.Q. Phenotypic and molecular detection of the bla KPC gene in clinical isolates from inpatients at hospitals in São Luis, MA, Brazil. BMC Infect. Dis. 2016, 16, 737. [Google Scholar] [CrossRef] [Green Version]
- Lima, W.G.; Silva Alves, G.C.; Sanches, C.; Antunes Fernandes, S.O.; de Paiva, M.C. Carbapenem-resistant Acinetobacter baumannii in patients with burn injury: A systematic review and meta-analysis. Burns 2019, 45, 1495–1508. [Google Scholar] [CrossRef]
- Martinez, T.; Martinez, I.; Vazquez, G.J.; Aquino, E.E.; Robledo, I.E. Genetic environment of the KPC gene in Acinetobacter baumannii ST2 clone from Puerto Rico and genomic insights into its drug resistance. J. Med. Microbiol. 2016, 65, 784–792. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zhou, Z.; Jiang, Y.; Yu, Y. Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 2011, 66, 1255–1259. [Google Scholar] [CrossRef] [PubMed]
- Jaidane, N.; Naas, T.; Oueslati, S.; Bernabeu, S.; Boujaafar, N.; Bouallegue, O.; Bonnin, R.A. Whole-genome sequencing of NDM-1-producing ST85 Acinetobacter baumannii isolates from Tunisia. Int. J. Antimicrob. Agents 2018, 52, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Beigverdi, R.; Sattari-Maraji, A.; Emaneini, M.; Jabalameli, F. Status of carbapenem-resistant Acinetobacter baumannii harboring carbapenemase: First systematic review and meta-analysis from Iran. Infect. Genet. Evol. 2019, 73, 433–443. [Google Scholar] [CrossRef]
- Salloum, T.; Tannous, E.; Alousi, S.; Arabaghian, H.; Rafei, R.; Hamze, M.; Tokajian, S. Genomic mapping of ST85 blaNDM-1 and blaOXA-94 producing Acinetobacter baumannii isolates from Syrian Civil War Victims. Int. J. Infect. Dis. 2018, 74, 100–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Mahdy, T.S.; Al-Agamy, M.H.; Al-Qahtani, A.A.; Shibl, A.M. Detection of blaOXA-23-like and blaNDM-1 in Acinetobacter baumannii from the Eastern Region, Saudi Arabia. Microb. Drug Resist. 2017, 23, 115–121. [Google Scholar] [CrossRef]
- Ramadan, R.A.; Gebriel, M.G.; Kadry, H.M.; Mosallem, A. Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: Characterization of carbapenemase genes and E-test evaluation of colistin-based combinations. Infect. Drug Resist. 2018, 11, 1261–1269. [Google Scholar] [CrossRef] [Green Version]
- Girija, S.A.; Jayaseelan, V.P.; Arumugam, P. Prevalence of VIM- and GIM-producing Acinetobacter baumannii from patients with severe urinary tract infection. Acta Microbiol. Immunol. Hung. 2018, 65, 539–550. [Google Scholar] [CrossRef] [Green Version]
- Gholami, M.; Moshiri, M.; Ahanjan, M.; Salimi Chirani, A.; Hasannejad Bibalan, M.; Asadi, A.; Eshaghi, M.; Pournajaf, A.; Abbasian, S.; Kouhsari, E.; et al. The diversity of class B and class D carbapenemases in clinical Acinetobacter baumannii isolates. Infez. Med. 2018, 26, 329–335. [Google Scholar]
- Aghamiri, S.; Amirmozafari, N.; Fallah Mehrabadi, J.; Fouladtan, B.; Hanafi Abdar, M. Antibiotic Resistance Patterns and a Survey of Metallo-β-Lactamase Genes Including bla-IMP and bla-VIM Types in Acinetobacter baumannii Isolated from Hospital Patients in Tehran. Chemotherapy 2016, 61, 275–280. [Google Scholar] [CrossRef]
- Rezaei, A.; Fazeli, H.; Halaji, M.; Moghadampour, M.; Faghri, J. Prevalence of metallo-beta-lactamase producing Acinetobacter baumannii isolated from intensive care unit in tertiary care hospitals. Ann. Ig. Med. Prev. Comunita 2018, 30, 330–336. [Google Scholar]
- Shakibaie, M.R.; Azizi, O.; Shahcheraghi, F. Insight into stereochemistry of a new IMP allelic variant (IMP-55) metallo-β-lactamase identified in a clinical strain of Acinetobacter baumannii. Infect. Genet. Evol. 2017, 51, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Leungtongkam, U.; Thummeepak, R.; Tasanapak, K.; Sitthisak, S. Acquisition and transfer of antibiotic resistance genes in association with conjugative plasmid or class 1 integrons of Acinetobacter baumannii. PLoS ONE 2018, 13, e0208468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.H.; Lee, J.H.; Lee, J.J.; Park, K.S.; Karim, A.M.; Lee, C.-R.; Jeong, B.C.; Lee, S.H. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int. J. Mol. Sci. 2015, 16, 9654–9692. [Google Scholar] [CrossRef]
- Héritier, C.; Poirel, L.; Nordmann, P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 2006, 12, 123–130. [Google Scholar] [CrossRef] [Green Version]
- Kumburu, H.H.; Sonda, T.; van Zwetselaar, M.; Leekitcharoenphon, P.; Lukjancenko, O.; Mmbaga, B.T.; Alifrangis, M.; Lund, O.; Aarestrup, F.M.; Kibiki, G.S. Using WGS to identify antibiotic resistance genes and predict antimicrobial resistance phenotypes in MDR Acinetobacter baumannii in Tanzania. J. Antimicrob. Chemother. 2019, 74, 1484–1493. [Google Scholar] [CrossRef]
- Jia, H.; Chen, Y.; Wang, J.; Ruan, Z. Genomic characterisation of a clinical Acinetobacter baumannii ST1928 isolate carrying a new ampC allelic variant blaADC-196 gene from China. J. Glob. Antimicrob. Resist. 2019, 19, 43–45. [Google Scholar] [CrossRef]
- Mayanskiy, N.; Chebotar, I.; Alyabieva, N.; Kryzhanovskaya, O.; Savinova, T.; Turenok, A.; Bocharova, Y.; Lazareva, A.; Polikarpova, S.; Karaseva, O. Emergence of the Uncommon Clone ST944/ST78 Carrying blaOXA-40-like and blaCTX-M-like Genes Among Carbapenem-Nonsusceptible Acinetobacter baumannii in Moscow, Russia. Microb. Drug Resist. 2017, 23, 864–870. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, X.; Zhou, Y.; Zhang, Z.; Zheng, X. Whole-genome sequencing of an NDM-1- and OXA-58-producing Acinetobacter towneri isolate from hospital sewage in Sichuan Province, China. J. Glob. Antimicrob. Resist. 2019, 16, 4–5. [Google Scholar] [CrossRef]
- Sarikhani, Z.; Nazari, R.; Nateghi Rostami, M. First report of OXA-143-lactamase producing Acinetobacter baumannii in Qom, Iran. Iran. J. Basic Med. Sci. 2017, 20, 1282–1286. [Google Scholar]
- Higgins, P.G.; Pérez-Llarena, F.J.; Zander, E.; Fernández, A.; Bou, G.; Seifert, H. OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013, 57, 2121–2126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paton, R.; Miles, R.S.; Hood, J.; Amyes, S.G.; Miles, R.S.; Amyes, S.G. ARI 1: Beta-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 1993, 2, 81–87. [Google Scholar] [CrossRef]
- Scaife, W.; Young, H.K.; Paton, R.H.; Amyes, S.G. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J. Antimicrob. Chemother. 1995, 36, 585–586. [Google Scholar] [CrossRef] [PubMed]
- Mugnier, P.D.; Poirel, L.; Naas, T.; Nordmann, P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 2010, 16, 35–40. [Google Scholar] [CrossRef]
- Figueiredo, S.; Poirel, L.; Papa, A.; Koulourida, V.; Nordmann, P. Overexpression of the naturally occurring blaOXA-51 gene in Acinetobacter baumannii mediated by novel insertion sequence ISAba9. Antimicrob. Agents Chemother. 2009, 53, 4045–4047. [Google Scholar] [CrossRef] [Green Version]
- Wibberg, D.; Salto, I.P.; Eikmeyer, F.G.; Maus, I.; Winkler, A.; Nordmann, P.; Pühler, A.; Poirel, L.; Schlüter, A. Complete Genome Sequencing of Acinetobacter baumannii Strain K50 Discloses the Large Conjugative Plasmid pK50a Encoding Carbapenemase OXA-23 and Extended-Spectrum β-Lactamase GES-11. Antimicrob. Agents Chemother. 2018, 62, e00212-18. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Patil, P.P.; Singhal, L.; Ray, P.; Patil, P.B.; Gautam, V. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates reveals the emergence of blaOXA-23 and blaNDM-1 encoding international clones in India. Infect. Genet. Evol. 2019, 75, 103986. [Google Scholar] [CrossRef]
- Mathlouthi, N.; Ben Lamine, Y.; Somai, R.; Bouhalila-Besbes, S.; Bakour, S.; Rolain, J.-M.; Chouchani, C. Incidence of OXA-23 and OXA-58 Carbapenemases Coexpressed in Clinical Isolates of Acinetobacter baumannii in Tunisia. Microb. Drug Resist. 2018, 24, 136–141. [Google Scholar] [CrossRef]
- Santimaleeworagun, W.; Samret, W.; Preechachuawong, P.; Kerdsin, A.; Jitwasinkul, T. Emergence of Co-Carbapenemase Genes, Bla(Oxa23), Bla(Vim) And Bla(Ndm) In Carbapenemresistant Acinetobacter baumannii Clinical Isolates. Southeast Asian J. Trop. Med. Public Health 2016, 47, 1001–1007. [Google Scholar]
- Zhu, L.-J.; Chen, X.-Y.; Hou, P.-F. Mutation of CarO participates in drug resistance in imipenem-resistant Acinetobacter baumannii. J. Clin. Lab. Anal. 2019, 33, e22976. [Google Scholar] [CrossRef] [Green Version]
- Benmahmod, A.B.; Said, H.S.; Ibrahim, R.H. Prevalence and Mechanisms of Carbapenem Resistance Among Acinetobacter baumannii Clinical Isolates in Egypt. Microb. Drug Resist. 2019, 25, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Cayô, R.; Rodríguez, M.-C.; Espinal, P.; Fernández-Cuenca, F.; Ocampo-Sosa, A.A.; Pascual, A.; Ayala, J.A.; Vila, J.; Martínez-Martínez, L. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 5907–5913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirshekar, M.; Shahcheraghi, F.; Azizi, O.; Solgi, H.; Badmasti, F. Diversity of Class 1 Integrons, and Disruption of carO and dacD by Insertion Sequences among Acinetobacter baumannii Isolates in Tehran, Iran. Microb. Drug Resist. 2018, 24, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Rumbo, C.; Gato, E.; López, M.; Ruiz de Alegría, C.; Fernández-Cuenca, F.; Martínez-Martínez, L.; Vila, J.; Pachón, J.; Cisneros, J.M.; Rodríguez-Baño, J.; et al. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013, 57, 5247–5257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Bilya, S.R.; Xu, W. adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 2019, 30, 100549. [Google Scholar] [CrossRef]
- Marchand, I.; Damier-Piolle, L.; Courvalin, P.; Lambert, T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 2004, 48, 3298–3304. [Google Scholar] [CrossRef] [Green Version]
- Dou, Q.; Zou, M.; Li, J.; Wang, H.; Hu, Y.; Liu, W. AdeABC efflux pump and resistance of Acinetobacter baumannii against carbapenem. Med. Sci. 2017, 42, 426–433. [Google Scholar]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. MMBR 2001, 65, 232–260. [Google Scholar] [CrossRef] [Green Version]
- Huys, G.; Cnockaert, M.; Vaneechoutte, M.; Woodford, N.; Nemec, A.; Dijkshoorn, L.; Swings, J. Distribution of tetracycline resistance genes in genotypically related and unrelated multiresistant Acinetobacter baumannii strains from different European hospitals. Res. Microbiol. 2005, 156, 348–355. [Google Scholar] [CrossRef]
- Magnet, S.; Courvalin, P.; Lambert, T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 2001, 45, 3375–3380. [Google Scholar] [CrossRef] [Green Version]
- Damier-Piolle, L.; Magnet, S.; Brémont, S.; Lambert, T.; Courvalin, P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2008, 52, 557–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuhan, Y.; Ziyun, Y.; Yongbo, Z.; Fuqiang, L.; Qinghua, Z. Over expression of AdeABC and AcrAB-TolC efflux systems confers tigecycline resistance in clinical isolates of Acinetobacter baumannii and Klebsiella pneumoniae. Rev. Soc. Bras. Med. Trop. 2016, 49, 165–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savari, M.; Ekrami, A.; Shoja, S.; Bahador, A. Plasmid borne Carbapenem-Hydrolyzing Class D β-Lactamases (CHDLs) and AdeABC efflux pump conferring carbapenem-tigecycline resistance among Acinetobacter baumannii isolates harboring TnAbaRs. Microb. Pathog. 2017, 104, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Costello, S.E.; Gales, A.C.; Morfin-Otero, R.; Jones, R.N.; Castanheira, M. Mechanisms of Resistance, Clonal Expansion, and Increasing Prevalence of Acinetobacter baumannii Strains Displaying Elevated Tigecycline MIC Values in Latin America. Microb. Drug Resist. 2016, 22, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M. Tigecycline: What is it, and where should it be used? J. Antimicrob. Chemother. 2005, 56, 611–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribera, A.; Ruiz, J.; Vila, J. Presence of the Tet M determinant in a clinical isolate of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2003, 47, 2310–2312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dönhöfer, A.; Franckenberg, S.; Wickles, S.; Berninghausen, O.; Beckmann, R.; Wilson, D.N. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 16900–16905. [Google Scholar] [CrossRef] [Green Version]
- Brown, P.C.; Borowska, E.; Schwartz, T.; Horn, H. Impact of the particulate matter from wastewater discharge on the abundance of antibiotic resistance genes and facultative pathogenic bacteria in downstream river sediments. Sci. Total Environ. 2019, 649, 1171–1178. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, M.; Zhong, X.; Liu, P.; Xie, X.; Wangxiao, J.; Sun, Y. Dissemination of resistance genes in duck/fish polyculture ponds in Guangdong Province: Correlations between Cu and Zn and antibiotic resistance genes. Environ. Sci. Pollut. Res. Int. 2019, 26, 8182–8193. [Google Scholar] [CrossRef]
- Perez, F.; Hujer, A.M.; Hujer, K.M.; Decker, B.K.; Rather, P.N.; Bonomo, R.A. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 3471–3484. [Google Scholar] [CrossRef] [Green Version]
- Hamouda, A.; Amyes, S.G.B. Novel gyrA and parC point mutations in two strains of Acinetobacter baumannii resistant to ciprofloxacin. J. Antimicrob. Chemother. 2004, 54, 695–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spence, R.P.; Towner, K.J. Frequencies and mechanisms of resistance to moxifloxacin in nosocomial isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 2003, 52, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Su, X.-Z.; Chen, J.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 2005, 49, 4362–4364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, Y.J.; Moon, D.C.; Jin, J.S.; Choi, C.H.; Lee, Y.C.; Lee, J.C. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn. Microbiol. Infect. Dis. 2009, 64, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Wachino, J.; Doi, Y.; Kurokawa, H.; Arakawa, Y. Global spread of multiple aminoglycoside resistance genes. Emerg. Infect. Dis. 2005, 11, 951–953. [Google Scholar] [CrossRef]
- Jana, S.; Deb, J.K. Molecular understanding of aminoglycoside action and resistance. Appl. Microbiol. Biotechnol. 2006, 70, 140–150. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, C.; Wu, J.; Jiang, R.; Mi, Z.; Huang, Z. A novel aminoglycoside-modifying enzyme gene aac(6′)-Ib in a pandrug-resistant Acinetobacter baumannii strain. J. Hosp. Infect. 2009, 73, 184–185. [Google Scholar] [CrossRef]
- Salimizand, H.; Zomorodi, A.R.; Mansury, D.; Khakshoor, M.; Azizi, O.; Khodaparast, S.; Baseri, Z.; Karami, P.; Zamanlou, S.; Farsiani, H.; et al. Diversity of aminoglycoside modifying enzymes and 16S rRNA methylases in Acinetobacter baumannii and Acinetobacter nosocomialis species in Iran; wide distribution of aadA1 and armA. Infect. Genet. Evol. 2018, 66, 195–199. [Google Scholar] [CrossRef]
- Hasani, A.; Sheikhalizadeh, V.; Ahangarzadeh Rezaee, M.; Rahmati-Yamchi, M.; Hasani, A.; Ghotaslou, R.; Goli, H.R. Frequency of Aminoglycoside-Modifying Enzymes and ArmA Among Different Sequence Groups of Acinetobacter baumannii in Iran. Microb. Drug Resist. 2016, 22, 347–353. [Google Scholar] [CrossRef]
- Doi, Y.; Adams, J.M.; Yamane, K.; Paterson, D.L. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob. Agents Chemother. 2007, 51, 4209–4210. [Google Scholar] [CrossRef] [Green Version]
- Sheikhalizadeh, V.; Hasani, A.; Ahangarzadeh Rezaee, M.; Rahmati-Yamchi, M.; Hasani, A.; Ghotaslou, R.; Goli, H.R. Comprehensive study to investigate the role of various aminoglycoside resistance mechanisms in clinical isolates of Acinetobacter baumannii. J. Infect. Chemother. 2017, 23, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Appleman, M.D.; Belzberg, H.; Citron, D.M.; Heseltine, P.N.; Yellin, A.E.; Murray, J.; Berne, T.V. In vitro activities of nontraditional antimicrobials against multiresistant Acinetobacter baumannii strains isolated in an intensive care unit outbreak. Antimicrob. Agents Chemother. 2000, 44, 1035–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, V.B.; Rajamohan, G.; Gebreyes, W.A. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 5312–5316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, U.; Yamashita, E.; Neuberger, A.; Morimoto, M.; van Veen, H.W.; Murakami, S. Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun. 2017, 8, 1336. [Google Scholar] [CrossRef] [PubMed]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-mediated mcr-1 gene in Acinetobacter baumannii and Pseudomonas aeruginosa: First report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, W.G.; Alves, M.C.; Cruz, W.S.; Paiva, M.C. Chromosomally encoded and plasmid-mediated polymyxins resistance in Acinetobacter baumannii: A huge public health threat. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1009–1019. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Boyce, J.D. Mechanisms of Polymyxin Resistance. Adv. Exp. Med. Biol. 2019, 1145, 55–71. [Google Scholar]
- Cheah, S.-E.; Johnson, M.D.; Zhu, Y.; Tsuji, B.T.; Forrest, A.; Bulitta, J.B.; Boyce, J.D.; Nation, R.L.; Li, J. Polymyxin Resistance in Acinetobacter baumannii: Genetic Mutations and Transcriptomic Changes in Response to Clinically Relevant Dosage Regimens. Sci. Rep. 2016, 6, 26233. [Google Scholar] [CrossRef] [Green Version]
- Trebosc, V.; Gartenmann, S.; Tötzl, M.; Lucchini, V.; Schellhorn, B.; Pieren, M.; Lociuro, S.; Gitzinger, M.; Tigges, M.; Bumann, D.; et al. Dissecting Colistin Resistance Mechanisms in Extensively Drug-Resistant Acinetobacter baumannii Clinical Isolates. mBio 2019, 10, e01083-19. [Google Scholar] [CrossRef] [Green Version]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.F.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, C.; Trent, M.S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014, 83, 99–128. [Google Scholar] [CrossRef] [PubMed]
- Hood, M.I.; Becker, K.W.; Roux, C.M.; Dunman, P.M.; Skaar, E.P. genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect. Immun. 2013, 81, 542–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Meletis, G.; Skoura, L. Polymyxin Resistance Mechanisms: From Intrinsic Resistance to Mcr Genes. Recent Pat. Anti-Infect. Drug Discov. 2018, 13, 198–206. [Google Scholar] [CrossRef]
- Martins-Sorenson, N.; Snesrud, E.; Xavier, D.E.; Cacci, L.C.; Iavarone, A.T.; McGann, P.; Riley, L.W.; Moreira, B.M. A novel plasmid-encoded mcr-4.3 gene in a colistin-resistant Acinetobacter baumannii clinical strain. J. Antimicrob. Chemother. 2020, 75, 60–64. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 119. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9030119
Ayoub Moubareck C, Hammoudi Halat D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics. 2020; 9(3):119. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9030119
Chicago/Turabian StyleAyoub Moubareck, Carole, and Dalal Hammoudi Halat. 2020. "Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen" Antibiotics 9, no. 3: 119. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9030119






