Human Defensins: A Novel Approach in the Fight against Skin Colonizing Staphylococcus aureus
Abstract
:1. Introduction
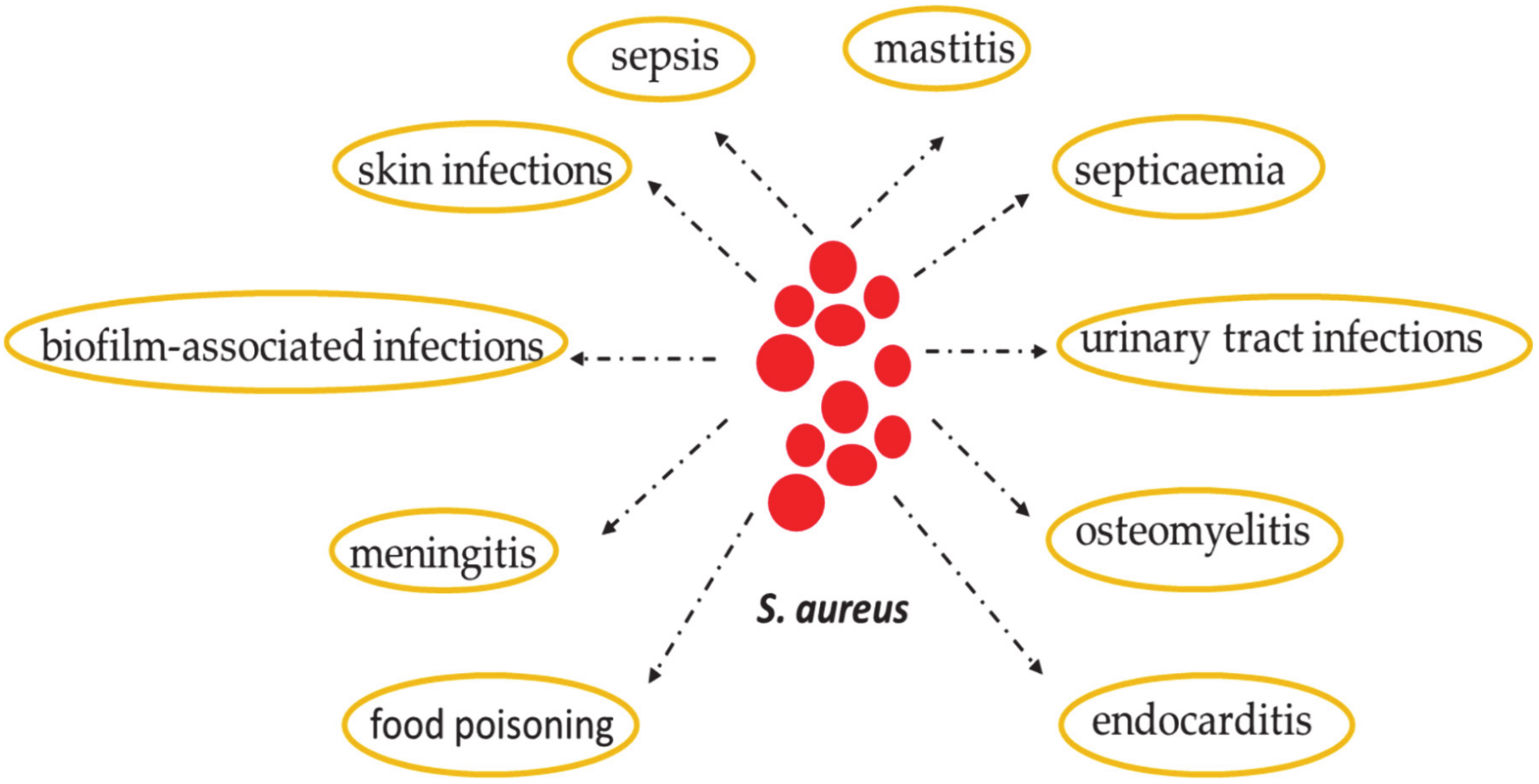
2. Staphylococcus aureus: Skin Colonization and Infection
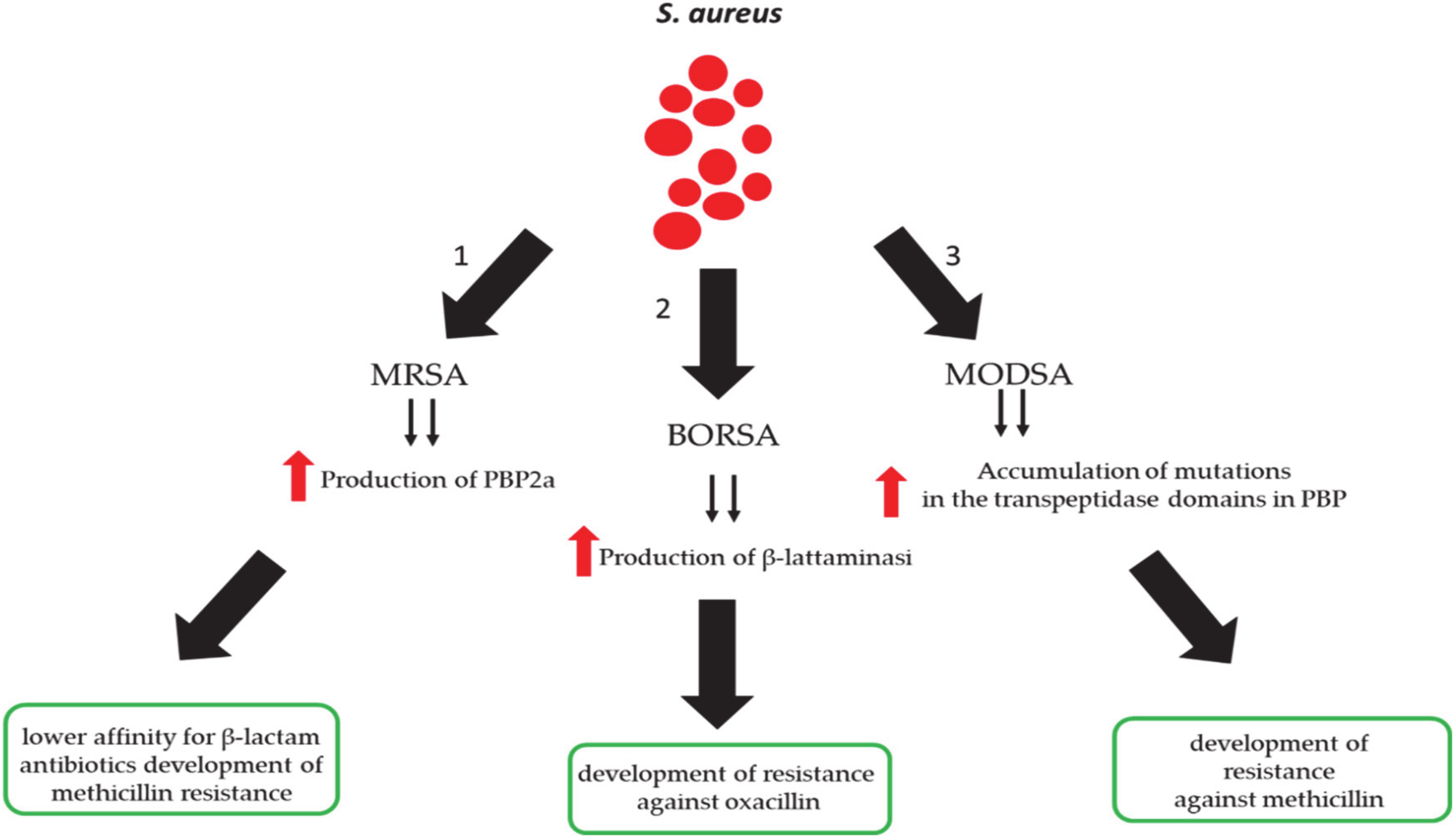
3. Methicillin-Resistant S. aureus (MRSA) Infection
4. Effective Human Defensins Versus S. aureus
4.1. Alfa-Defensins
4.2. Beta-Defensins
5. Antimicrobial Barrier of the Skin
6. Human Defensins and S. aureus-Dependent Skin Diseases
7. Common S. aureus Cutaneous Infections
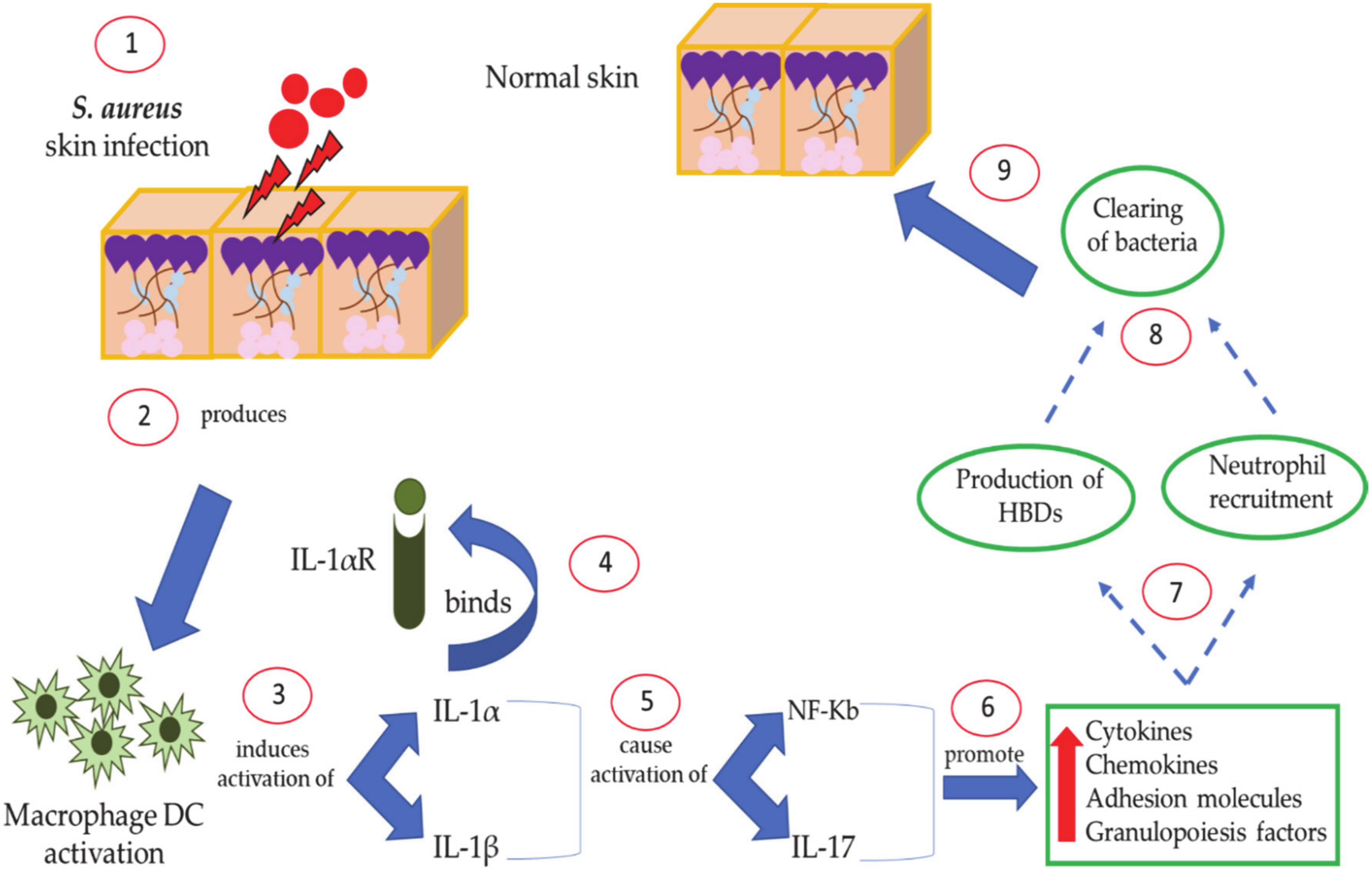
8. Host Defense Mediated by Human Defenses against S. aureus Skin Disease: The Role of IL-1 and IL-17
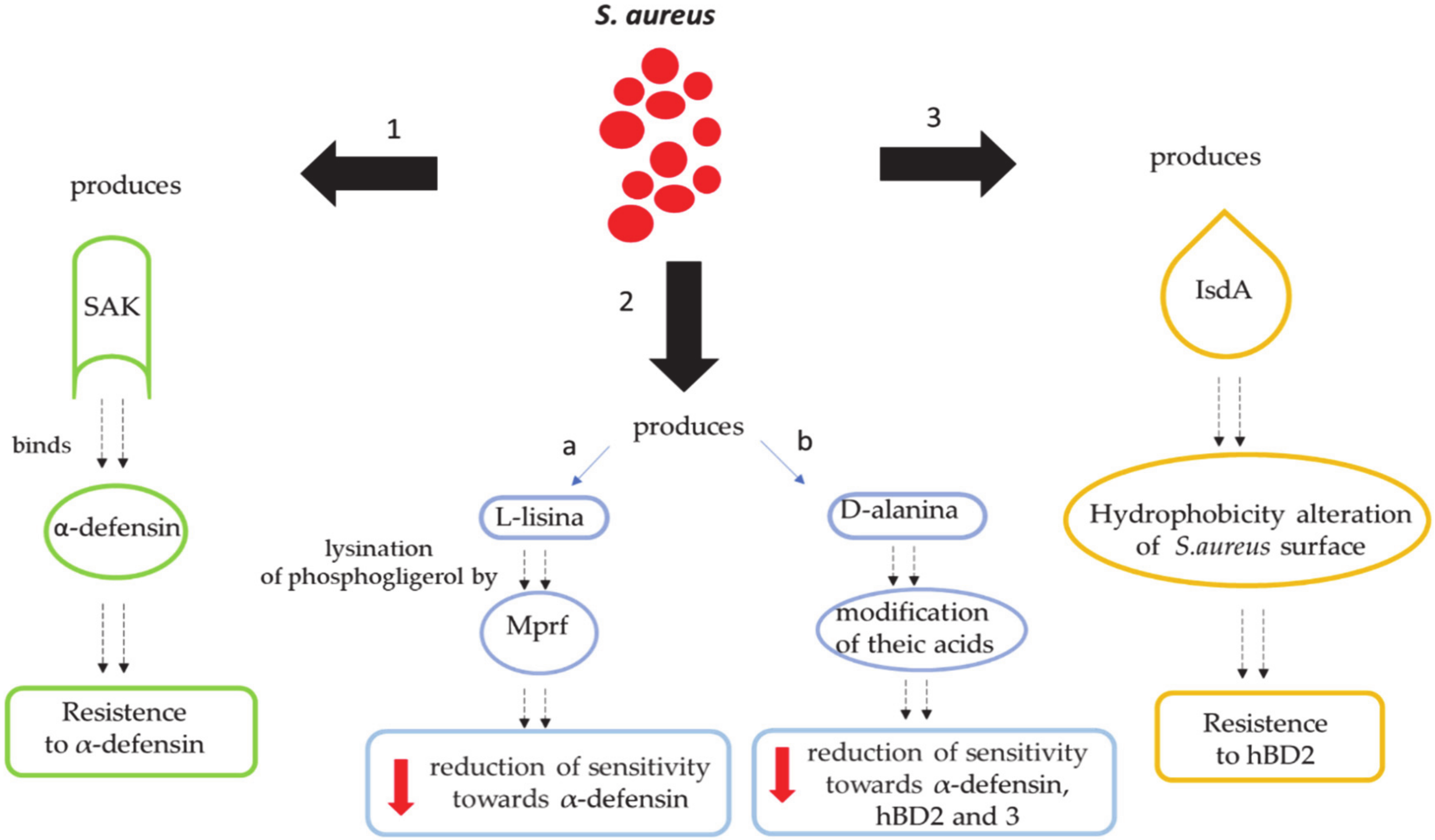
9. Mechanisms by Which S. aureus Escapes the Human Defenses Derived from the Skin
9.1. Secretion of Molecules Binding Extracellular Defensins
9.2. Reduction of Net Negative Charges on the Bacterial Surface
9.2.1. S. aureus’ Alteration of the Cell Membrane with L-lysine
9.2.2. S. aureus’ Alteration of the Cell Membrane with D-Alanine
9.3. Modification of Skin Hydrophobicity
10. Conclusions and Future Perspective
Funding
Conflicts of Interest
References
- van Wamel, W.J.B. Staphylococcus aureus infections, some second thoughts. Curr. Opin. Infect. Dis. 2017, 30, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K. Staphylococcus aureus intracellular survival: A closer look in the process. Virulence 2017, 8, 1506–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Querques, F.; Cantilena, B.; Cozzolino, C.; Esposito, M.T.; Passaro, F.; Parisi, S.; Lombardo, B.; Russo, T.; Pastore, L. Angiotensin receptor I stimulates osteoprogenitor proliferation through TGFβ-mediated signaling. J. Cell. Physiol. 2015, 23, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Asgeirsson, H.; Thalme, A.; Weiland, O. Staphylococcus aureus bacteraemia and endocarditis—epidemiology and outcome: A review. Infect. Dis. 2018, 3, 175–192. [Google Scholar] [CrossRef] [PubMed]
- Saeed, K.; Bal, A.M.; Gould, I.M.; David, M.Z.; Dryden, M.; Giannitsioti, E.; Hijazi, K.; Meisner, J.A.; Esposito, S.; Scaglione, F.; et al. An update on Staphylococcus aureus infective endocarditis from the International Society of Antimicrobial Chemotherapy (ISAC). Int. J. Antimicrob. Agents 2019, 53, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Pletz, M.W.; Burkhardt, O.; Welte, T. Nosocomial methicillin-resistant Staphylococcus aureus (MRSA) pneumonia: Linezolid or vancomycin?—Comparison of pharmacology and clinical efficacy. Eur. J. Med. Res. 2010, 15, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Lesher, B.; Gao, X.; Chen, Y.; Liu, Z. Methicillin-resistant Staphylococcus aureus nosocomial pneumonia: Role of linezolid in the People’s Republic of China. Clin. Outcomes Res. 2016, 8, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Shi, B.; Leung, D.Y.; Taylor, P.A.; Li, H. Methicillin-resistant Staphylococcus aureus colonization is associated with decreased skin commensal bacteria in atopic dermatitis. J. Invest. Dermatol. 2018, 138, 1668–1671. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kim, B.E.; Leung, D.Y. Pathophysiology of atopic dermatitis: Clinical implications. Allergy Asthma Proc. 2019, 40, 84–92. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, M.E.; Wardenburg, J.B. Igniting the Fire: Staphylococcus aureus Virulence Factors in the Pathogenesis of Sepsis. PLoS Pathog. 2014, 10, e1003871. [Google Scholar] [CrossRef] [PubMed]
- Archer, N.K.; Mazaitis, M.J.; William Costerton, J.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms: Properties, regulation and roles in human disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Paterson, G.K.; Harrison, E.M.; Holmes, M.A. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014, 22, 42–47. [Google Scholar] [CrossRef] [Green Version]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2019, 95, 241. [Google Scholar] [CrossRef]
- Erskine, R.J. Vaccination Strategies for Mastitis. Vet. Clin. N. Am. Food Anim. Pract. 2012, 28, 257–270. [Google Scholar] [CrossRef]
- Leitner, G.; Lubashevsky, E.; Glickman, A.; Winkler, M.; Saran, A.; Trainin, Z. Development of a Staphylococcus aureus vaccine against mastitis in dairy cows: I. Challenge trials. Vet. Immunol. Immunopathol. 2003, 93, 31–38. [Google Scholar] [CrossRef]
- Keefe, G. Update on control of Staphylococcus aureus and Streptococcus agalactiae for management of mastitis. Vet. Clin. N. Am. Food Anim. Pract. 2012, 28, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Heringstad, B.; Klemetsdal, G.; Ruane, J. Selection for mastitis resistance in dairy cattle: A review with focus on the situation in the Nordic countries. Livest. Prod. Sci. 2000, 64, 95–106. [Google Scholar] [CrossRef]
- Rupp, R.; Boichard, D. Genetics of resistance to mastitis in dairy cattle. Vet. Res. 2003, 34, 671–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bangert, C.; Brunner, P.M.; Stingl, G. Immune functions of the skin. Clin. Dermatol. 2011, 29, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Song, P.I.; Seo, C.H.; Cheong, H.; Park, Y. Colonization and infection of the skin by S. aureus: immune system evasion and the response to cationic antimicrobial peptides. Int. J. Mol. Sci. 2014, 15, 8753–8772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorwitz, R.J.; Kruszon-Moran, D.; McAllister, S.K.; McQuillan, G.; McDougal, L.K.; Fosheim, G.E.; Jensen, B.J.; Killgore, G.; Tenover, F.C.; Kuehnert, M.J. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J. Infect. Dis. 2008, 197, 1226–1234. [Google Scholar] [CrossRef] [Green Version]
- Miller, L.G.; Diep, B.A. Clinical practice: Colonization, fomites, and virulence: Rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 2008, 46, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Program, N.C.S.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [Green Version]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Miajlovic, H.; Fallon, P.G.; Irvine, A.D.; Foster, T.J. Effect of filaggrin breakdown products on growth of and protein expression by Staphylococcus aureus. J. Allergy Clin. Immunol. 2010, 126, 1184–1190. [Google Scholar] [CrossRef] [Green Version]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef]
- Cogen, A.L.; Yamasaki, K.; Sanchez, K.M.; Dorschner, R.A.; Lai, Y.; MacLeod, D.T.; Torpey, J.W.; Otto, M.; Nizet, V.; Kim, J.E.; et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 2010, 130, 192–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, Y.; Cogen, A.L.; Radek, K.A.; Park, H.J.; Macleod, D.T.; Leichtle, A.; Ryan, A.F.; Di Nardo, A.; Gallo, R.L. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Invest. Dermatol. 2010, 130, 2211–2221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Gotz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J. Invest. Dermatol. 2011, 131, 382–390. [Google Scholar] [CrossRef] [Green Version]
- Braff, M.H.; Zaiou, M.; Fierer, J.; Nizet, V.; Gallo, R.L. Keratinocyte production of cathelicidin provides direct activity against bacterial skin pathogens. Infect. Immun. 2005, 73, 6771–6781. [Google Scholar] [CrossRef] [Green Version]
- Herman, A.; Herman, A.P. Antimicrobial peptides activity in the skin. Skin Res Technol. 2019, 25, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Kisich, K.O.; Howell, M.D.; Boguniewicz, M.; Heizer, H.R.; Watson, N.U.; Leung, D.Y. The constitutive capacity of human keratinocytes to kill Staphylococcus aureus is dependent on β-defensin 3. J. Invest. Dermatol. 2007, 127, 2368–2380. [Google Scholar] [CrossRef] [Green Version]
- Simanski, M.; Dressel, S.; Glaser, R.; Harder, J. RNase 7 protects healthy skin from Staphylococcus aureus colonization. J. Invest. Dermatol. 2010, 130, 2836–2838. [Google Scholar] [CrossRef] [Green Version]
- Ong, P.Y.; Ohtake, T.; Brandt, C.; Strickland, I.; Boguniewicz, M.; Ganz, T.; Gallo, R.L.; Leung, D.Y. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 2002, 347, 1151–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burian, M.; Rautenberg, M.; Kohler, T.; Fritz, M.; Krismer, B.; Unger, C.; Hoffmann, W.H.; Peschel, A.; Wolz, C.; Goerke, C. Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 2010, 201, 1414–1421. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.H.; Strickland, I.; Tomkinson, A.; Fehringer, A.P.; Gelfand, E.W.; Leung, D.Y. Preferential binding of Staphylococcus aureus to skin sites of Th2-mediated inflammation in a murine model. J. Invest. Dermatol. 2001, 116, 658–663. [Google Scholar] [CrossRef] [Green Version]
- Clarke, S.R.; Brummell, K.J.; Horsburgh, M.J.; McDowell, P.W.; Mohamad, S.A.; Stapleton, M.R.; Acevedo, J.; Read, R.C.; Day, N.P.; Peacock, S.J.; et al. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J. Infect. Dis. 2006, 193, 1098–1108. [Google Scholar] [CrossRef] [Green Version]
- Weidenmaier, C.; Kokai-Kun, J.F.; Kristian, S.A.; Chanturiya, T.; Kalbacher, H.; Gross, M.; Nicholson, G.; Neumeister, B.; Mond, J.J.; Peschel, A. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 2004, 10, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Laouini, D.; Kawamoto, S.; Yalcindag, A.; Bryce, P.; Mizoguchi, E.; Oettgen, H.; Geha, R.S. Epicutaneous sensitization with superantigen induces allergic skin inflammation. J. Allergy. Clin. Immunol. 2003, 112, 981–987. [Google Scholar] [CrossRef]
- Clarke, S.R.; Mohamed, R.; Bian, L.; Routh, A.F.; Kokai-Kun, J.F.; Mond, J.J.; Tarkowski, A.; Foster, S.J. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 2007, 1, 199–212. [Google Scholar] [CrossRef] [Green Version]
- Aguayo-Reyes, A.; Quezada-Aguiluz, M.; Mella, S.; Riedel, G.; Opazo-Capurro, A.; Bello-Toledo, H.; Domínguez, M.; González-Rocha, G. Bases moleculares de la resistencia a meticilina en Staphylococcus aureus. Rev. Chil. Infectol. 2018, 35, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, P.D.; Taylor, P.W. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog. 2002, 85, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Tsubakishita, S.; Kuwahara-Arai, K.; Baba, T.; Hiramatsu, K. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob. Agents Chemother. 2010, 54, 1469–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Alvarez, L.; Holden, M.T.G.; Lindsay, H.; Webb, C.R.; Brown, D.F.J.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: Adescriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef] [Green Version]
- MacFadyen, A.C.; Fisher, E.A.; Costa, B.; Cullen, C.; Paterson, G.K. Genome analysis of methicillin resistance in Macrococcus caseolyticus from dairy cattle in England and Wales. Microb. Genom. 2018, 4, 1–8. [Google Scholar] [CrossRef]
- Argudín, M.A.; Roisin, S.; Nienhaus, L.; Dodémont, M.; de Mendonça, R.; Nonhoff, C.; Deplano, A.; Denis, O. Genetic diversity among Staphylococcus aureus isolates showing oxacillin and/or cefoxitin resistance not linked to the presence of mec genes. Antimicrob. Agents Chemother. 2018, 62, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiemersma, E.W.; Bronzwaer, S.L.; Lyytikäinen, O.; Degener, J.E.; Schrijnemakers, P.; Bruinsma, N.; Monen, J.; Witte, W.; Grundman, H. European Antimicrobial Resistance Surveillance System Participants. Methicillin-resistant Staphylococcus aureus in Europe, 1999–2002. Emerg. Infect. Dis. 2004, 10, 1627–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, A.D.; Otto, M.; Braughton, K.R.; Whitney, A.R.; Chen, L.; Mathema, B. Epidemic community-associated methicillin-resistant Staphylococcus aureus: Recent clonal expansionand diversification. Proc. Natl. Acad. Sci. USA 2008, 105, 1327–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golding, G.R.; Campbell, J.L.; Spreitzer, D.J.; Veyhl, J.; Surynicz, K.; Simor, A. A preliminaryguideline for the assignment of methicillin-resistant Staphylococcus aureus to a Canadian pulsed-field gel electrophoresis epidemic type using spa typing. Can. J. Infect. Dis. Med. Microbiol. 2008, 19, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.X.; Ito, T.; Tiensasitorn, C.; Jamklang, M.; Chongtrakool, P.; Boyle-Vavra, S.; Daum, R.S.; Hiramatsu, K. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2002, 46, 1147–1152. [Google Scholar] [CrossRef] [Green Version]
- Pozzi, C.; Waters, E.M.; Rudkin, J.K.; Schaeffer, C.R.; Lohan, A.J.; Tong, P.; Loftus, B.J.; Pier, G.B.; Fay, P.D.; Massey, R.C.; et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012, 8, e1002626. [Google Scholar] [CrossRef]
- Berglund, C.; Söderquist, B. The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden—Possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus. Clin. Microbiol. Infect. 2008, 14, 1048–1056. [Google Scholar] [CrossRef] [Green Version]
- Hososaka, Y.; Hanaki, H.; Endo, H.; Suzuki, Y.; Nagasawa, Z.; Otsuka, Y.; Nakae, T.; Sunakawa, K. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: A new type of MRSA. J. Infect. Chemother. 2007, 13, 79–86. [Google Scholar] [CrossRef]
- Belcheva, A.; Golemi-Kotra, D.A. Close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J. Biol. Chem. 2008, 283, 12354–12364. [Google Scholar] [CrossRef] [Green Version]
- Boyle-Vavra, S.; Yin, S.; Daum, R.S. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 2006, 262, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Chuang, Y.Y.; Huang, Y.C. Livestock-associated methicillin-resistant Staphylococcus aureus in Asia: An emerging issue? Int. J. Antimicrob. Agents 2015, 45, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Molla, B.; Byrne, M.; Abley, M.; Mathews, J.; Jackson, C.R.; Fedorka-Cray, P.; Sreevatsan, S.; Wang, P.; Gebreyes, W.A. Epidemiology and genotypic characteristics of methicillin resistant Staphylococcus aureus strains of porcine origin. J. Clin. Microbiol. 2012, 50, 3687–3693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundmann, H.; Schouls, L.M.; Aanensen, D.M.; Pluister, G.N.; Tami, A.; Chlebowicz, M.; Glasner, C.; Sabat, A.J.; Weist, K.; Heuer, O.; et al. ESCMID Study Group on Molecular Epidemiological Markers; European Staphylococcal Reference Laboratory Working Group. The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. Euro Surveill. 2014, 19, 20987. [Google Scholar] [PubMed]
- Cho, J.S.; Xuan, C.; Miller, L.S. Lucky number seven: RNase 7 can prevent Staphylococcus aureus skin colonization. J. Invest. Dermatol. 2010, 130, 2703–2706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev. Dermatol. 2010, 5, 183–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallo, R.L.; Huttner, K.M. Antimicrobial peptides: An emerging concept in cutaneous biology. J. Invest. Dermatol. 1998, 111, 739–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahl, H.G.; Pag, U.; Bonness, S.; Wagner, S.; Antcheva, N.; Tossi, A. Mammalian defensins: Structures and mechanism of antibiotic activity. J. Leukoc. Biol. 2005, 77, 466–475. [Google Scholar] [CrossRef]
- Miller, L.S.; Sorensen, O.E.; Liu, P.T.; Jalian, H.R.; Eshtiaghpour, D.; Behmanesh, B.E.; Chung, W.; Starner, T.D.; Kim, J.; Sieling, P.A.; et al. TGF-α regulates TLR expression and function on epidermal keratinocytes. J. Immunol. 2005, 174, 6137–6143. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, O.E.; Thapa, D.R.; Roupe, K.M.; Valore, E.V.; Sjobring, U.; Roberts, A.A.; Schmidtchen, A.; Ganz, T. Injury-induced innate immune response in human skin mediated by transactivation of the epidermal growth factor receptor. J. Clin. Invest. 2006, 116, 1878–1885. [Google Scholar] [CrossRef] [Green Version]
- Grigat, J.; Soruri, A.; Forssmann, U.; Riggert, J.; Zwirner, J. Chemoattraction of macrophages, T lymphocytes, and mast cells is evolutionarily conserved within the human α-defensin family. J. Immunol. 2007, 179, 3958–3965. [Google Scholar] [CrossRef] [Green Version]
- Rohrl, J.; Yang, D.; Oppenheim, J.J.; Hehlgans, T. Human β -defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 2010, 184, 6688–6694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Chertov, O.; Bykovskaia, S.N.; Chen, Q.; Buffo, M.J.; Shogan, J.; Anderson, M.; Schroder, J.M.; Wang, J.M.; Howard, O.M.; et al. β-Defensins: Linking innate and adaptive immunity through dendritic and T cell CCR6. Science 1999, 286, 525–528. [Google Scholar] [CrossRef]
- Lehrer, R.I. Multispecific myeloid defensins. Curr. Opin. Hematol. 2007, 14, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Ericksen, B.; Wu, Z.; Lu, W.; Lehrer, R.I. Antibacterial activity and specificity of the six human α-defensins. Antimicrob. Agents Chemother. 2005, 49, 269–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Schauber, J.; Gallo, R.L. Antimicrobial peptides and the skin immune defense system. J. Allergy. Clin. Immunol. 2009, 124, R13–R18. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.; Meyer-Hoffert, U.; Wehkamp, K.; Schwichtenberg, L.; Schroder, J.M. Differential gene induction of human β-defensins (hBD-1, -2, -3, and -4) in keratinocytes is inhibited by retinoic acid. J. Invest. Dermatol. 2004, 123, 522–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harder, J.; Bartels, J.; Christophers, E.; Schroder, J.M. A peptide antibiotic from human skin. Nature 1997, 387, 861. [Google Scholar] [CrossRef]
- Garcia, J.R.; Krause, A.; Schulz, S.; Rodriguez-Jimenez, F.J.; Kluver, E.; Adermann, K.; Forssmann, U.; Frimpong-Boateng, A.; Bals, R.; Forssmann, W.G. Human β-defensin 4: A novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 2001, 15, 1819–1821. [Google Scholar] [CrossRef] [Green Version]
- Harder, J.; Bartels, J.; Christophers, E.; Schroder, J.M. Isolation and characterization of human β-defensin 3, a novel human inducible peptide antibiotic. J. Biol. Chem. 2001, 276, 5707–5713. [Google Scholar] [CrossRef] [Green Version]
- Dinulos, J.G.; Mentele, L.; Fredericks, L.P.; Dale, B.A.; Darmstadt, G.L. Keratinocyte expression of human β-defensin 2 following bacterial infection: Role in cutaneous host defense. Clin. Diagn. Lab. Immunol. 2003, 10, 161–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayama, K.; Komatsuzawa, H.; Yamasaki, K.; Shirakata, Y.; Hanakawa, Y.; Ouhara, K.; Tokumaru, S.; Dai, X.; Tohyama, M.; Ten Dijke, P.; et al. New mechanisms of skin innate immunity: ASK1-mediated keratinocyte differentiation regulates the expression of β-defensins, LL37, and TLR2. Eur. J. Immunol. 2005, 35, 1886–1895. [Google Scholar] [CrossRef]
- Menzies, B.E.; Kenoyer, A. Signal transduction and nuclear responses in Staphylococcus aureus-induced expression of human β-defensin 3 in skin keratinocytes. Infect. Immun. 2006, 74, 6847–6854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumikawa, Y.; Asada, H.; Hoshino, K.; Azukizawa, H.; Katayama, I.; Akira, S.; Itami, S. Induction of β-defensin 3 in keratinocytes stimulated by bacterial lipopeptides through Toll-like receptor 2. Microbes Infect. 2006, 8, 1513–1521. [Google Scholar] [CrossRef]
- Zanger, P.; Holzer, J.; Schleucher, R.; Scherbaum, H.; Schittek, B.; Gabrysch, S. Severity of Staphylococcus aureus infection of the skin is associated with inducibility of human β-defensin 3 but not human β-defensin 2. Infect. Immun. 2010, 78, 3112–3117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastore, S.; Mascia, F.; Mariani, V.; Girolomoni, G. The epidermal growth factor receptor system in skin repair and inflammation. J. Invest. Dermatol. 2008, 128, 1365–1374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, L.S.; Modlin, R.L. Human keratinocyte Toll-like receptors promote distinct immune responses. J. Invest. Dermatol. 2007, 127, 262–263. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Chertov, O.; Oppenheim, J.J. Participation of mammalian defensins and cathelicidins in anti-microbial immunity: Receptors and activities of human defensins and cathelicidin (LL-37). J. Leukoc. Biol. 2001, 69, 691–697. [Google Scholar]
- Bird, J.A.; Sánchez-Borges, M.; Ansotegui, I.J.; Ebisawa, M.; Ortega Martell, J.A. Skin as an immune organ and clinical applications of skin-based immunotherapy. World Allergy Organ J. 2018, 11, 38. [Google Scholar] [CrossRef] [Green Version]
- Nestle, F.O.; Di Meglio, P.; Qin, J.Z.; Nickoloff, B.J. Skin immune sentinels in health and disease. Nat. Rev. Immunol. 2009, 9, 679–691. [Google Scholar] [CrossRef] [Green Version]
- Krishna, S.; Miller, L.S. Innate and adaptive immune responses against Staphylococcus aureus skin infections. Semin. Immunopathol. 2012, 34, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Gallo, R.L. Toll-like receptors in skin infections and inflammatory diseases. Infect. Disord. Drug Targets 2008, 8, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Gallo, R.L. AMPed up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, L.S. Toll-like receptors in skin. Adv. Dermatol. 2008, 24, 71–87. [Google Scholar] [CrossRef] [Green Version]
- Rivas-Santiago, B.; Trujillo, V.; Montoya, A.; Gonzalez-Curiel, I.; Castaneda-Delgado, J.; Cardenas, A.; Rincon, K.; Hernandez, M.L.; Hernández-Pando, R. Expression of antimicrobial peptides in diabetic foot ulcer. J. Dermatol. Sci. 2012, 65, 19–26. [Google Scholar] [CrossRef]
- Lande, R.; Chamilos, G.; Ganguly, D.; Demaria, O.; Frasca, L.; Durr, S.; Conrad, C.; Schröder, J.; Gilliet, M. Cationic antimicrobial peptides in psoriatic skin cooperate to break innate tolerance to self-DNA. Eur. J. Immunol. 2015, 45, 203–213. [Google Scholar] [CrossRef]
- Takahashi, T.; Gallo, R.L. The Critical and Multifunctional Roles of Antimicrobial Peptides in Dermatology. Dermatol. Clin. 2017, 35, 39–50. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020. [Google Scholar] [CrossRef]
- Niyonsaba, F.; Kiatsurayanon, C.; Chieosilapatham, P.; Ogawa, H. Friends or Foes? Host defense (antimicrobial) peptides and proteins in human skin diseases. Exp. Dermatol. 2017, 26, 989–998. [Google Scholar] [CrossRef] [Green Version]
- Kiatsurayanon, C.; Niyonsaba, F.; Smithrithee, R.; Akiyama, T.; Ushio, H.; Hara, M.; Okumura, K.; Ikeda, S.; Ogawa, H. Host defense (Antimicrobial) peptide, human β-defensin-3, improves the function of the epithelial tight-junction barrier in human keratinocytes. J. Invest. Dermatol. 2014, 134, 2163–2173. [Google Scholar] [CrossRef] [Green Version]
- Korting, H.C.; Schöllmann, C.; Stauss-Grabo, M.; Schäfer-Korting, M. Antimicrobial peptides and skin: A paradigm of translational medicine. Skin Pharmacol. Physiol. 2012, 25, 323–334. [Google Scholar] [CrossRef] [Green Version]
- Midorikawa, K.; Ouhara, K.; Komatsuzawa, H.; Kawai, T.; Yamada, S.; Fujiwara, T.; Yamazaki, K.; Sayama, K.; Taubman, M.A.; Kurihara, H.; et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect. Immun. 2003, 71, 3730–3739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Niyonsaba, F.; Ushio, H.; Okuda, D.; Nagaoka, I.; Ikeda, S.; Okumura, K.; Ogawa, H. Synergistic effect of antibacterial agents human beta-defensins, cathelicidin LL-37 and lysozyme against Staphylococcus aureus and Escherichia coli. J. Dermatol. Sci. 2005, 40, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Sass, V.; Pag, U.; Tossi, A.; Bierbaum, G.; Sahl, H.G. Mode of action of human beta-defensin 3 against Staphylococcus aureus and transcriptional analysis of responses to defensin challenge. Int. J. Med. Microbiol. 2008, 298, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Kisich, K.O.; Carspecken, C.W.; Fiéve, S.; Boguniewicz, M.; Leung, D.Y. Defective killing of Staphylococcus aureus in atopic dermatitis is associated with reduced mobilization of human beta-defensin-3. J. Allergy Clin. Immunol. 2008, 122, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Mysliwy, J.; Spudy, B.; Lorenzen, I.; Reiss, K.; Gelhaus, C.; Podschun, R.; Leippe, M.; Grötzinger, J. Human beta-defensin 2 and beta-defensin 3 chimeric peptides reveal the structural basis of the pathogen specificity of their parent molecules. Antimicrob. Agents Chemother. 2011, 55, 954–960. [Google Scholar] [CrossRef] [Green Version]
- Zanger, P.; Nurjadi, D.; Vath, B.; Kremsner, P.G. Persistent nasal carriage of Staphylococcus aureus is associated with deficient induction of human beta-defensin 3 after sterile wounding of healthy skin in vivo. Infect. Immun. 2011, 79, 2658–2662. [Google Scholar] [CrossRef] [Green Version]
- Linzmeier, R.; Ho, C.H.; Hoang, B.V.; Ganz, T. A 450-kb contig of defensin genes on human chromosome 8p23. Gene 1999, 11, 205–211. [Google Scholar] [CrossRef]
- van Belkum, A.; Emonts, M.; Wertheim, H.; de Jongh, C.; Nouwen, J.; Bartels, H.; Cole, A.; Cole, A.; Hermans, P.; Boelens, H.; et al. The role of human innate immune factors in nasal colonization by Staphylococcus aureus. Microbes Infect. 2007, 9, 1471–1477. [Google Scholar] [CrossRef]
- Fode, P.; Stegger, M.; Andersen, P.S. Uman β-defensin 3 (DEFB103) and its influence on Staphylococcus aureus nasal carriage. Int. J. Infect. Dis. 2011, 15, e388–e394. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, K.; Gallo, R.L. Antimicrobial peptides in human skin disease. Eur. J. Dermatol. 2008, 18, 11–21. [Google Scholar] [PubMed]
- Eckmann, C.; Dryden, M. Treatment of complicated skin and soft-tissue infections caused by resistant bacteria: value of linezolid, tigecycline, daptomycin and vancomycin. Eur. J. Med. Res. 2010, 15, 554–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amara, S.; Adamson, R.T.; Lew, I.; Huang, X. Clinical response at Day 3 of therapy and economic outcomes in hospitalized patients with acute bacterial skin and skin structure infection (ABSSSI). Curr. Med. Res. Opin. 2013, 29, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Thangamani, S.; Younis, W.; Seleem, M.N. Repurposing ebselen for treatment of multidrug-resistant staphylococcal infections. Sci. Rep. 2015, 5, 11596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillamet, C.V.; Vazquez, R.; Noe, J.; Micek, S.T.; Kollef, M.H. A cohort study of bacteremic pneumonia: The importance of antibiotic resistance and appropriate initial therapy? Medicine 2016, 95, e4708. [Google Scholar] [CrossRef]
- Cardona, A.F.; Wilson, S.E. Skin and Soft-Tissue Infections: A Critical Review and the Role of Telavancin in Their Treatment. Clin. Infect. Dis. 2015, 61, S69–S78. [Google Scholar] [CrossRef] [Green Version]
- Esposito, S.; Bassetti, M.; Concia, E.; De Simone, G.; De Rosa, F.G.; Grossi, P.; Novelli, A.; Menichetti, F.; Petrosillo, N.; Tinelli, M.; et al. Italian Society of Infectious and Tropical Diseases. J. Chemother. 2017, 29, 197–214. [Google Scholar] [CrossRef]
- Stein, G.E.; Wells, E.M. The importance of tissue penetration in achieving successful antimicrobial treatment of nosocomial pneumonia and complicated skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus: vancomycin and linezolid. Curr. Med. Res. Opin. 2010, 26, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.F.; Hamed, M.I.; Panitch, A.; Seleem, M.N. Targeting methicillin-resistant Staphylococcus aureus with short salt-resistant synthetic peptides. Antimicrob. Agents Chemother. 2014, 58, 4113–4122. [Google Scholar] [CrossRef] [Green Version]
- Miller, L.S.; Cho, J.S. Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 2011, 11, 505–518. [Google Scholar] [CrossRef] [Green Version]
- Cua, D.J.; Tato, C.M. Innate IL-17-producing cells: The sentinels of the immune system. Nat. Rev. Immunol. 2010, 10, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Bokarewa, M.; Foster, T.; Mitchell, J.; Higgins, J.; Tarkowski, A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 2004, 172, 1169–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuhaus, F.C.; Baddiley, J. A continuum of anionic charge: Structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staubitz, P.; Neumann, H.; Schneider, T.; Wiedemann, I.; Peschel, A. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 2004, 231, 67–71. [Google Scholar] [CrossRef] [Green Version]
- Oku, Y.; Kurokawa, K.; Ichihashi, N.; Sekimizu, K. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 2004, 150, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Jann, N.J.; Schmaler, M.; Kristian, S.A.; Radek, K.A.; Gallo, R.L.; Nizet, V.; Peschel, A.; Landmann, R. Neutrophil antimicrobial defense against Staphylococcus aureus is mediated by phagolysosomal but not extracellular trap-associated cathelicidin. J. Leukoc. Biol. 2009, 86, 1159–1169. [Google Scholar] [CrossRef] [Green Version]
- Peschel, A.; Otto, M.; Jack, R.W.; Kalbacher, H.; Jung, G.; Gotz, F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999, 274, 8405–8410. [Google Scholar] [CrossRef] [Green Version]
- Simanski, M.; Glaser, R.; Koten, B.; Meyer-Hoffert, U.; Wanner, S.; Weidenmaier, C.; Peschel, A.; Harder, J. Staphylococcus aureus subverts cutaneous defense by D-alanylation of teichoic acids. Exp. Dermatol. 2013, 22, 294–296. [Google Scholar] [CrossRef]
- Neumann, Y.; Ohlsen, K.; Donat, S.; Engelmann, S.; Kusch, H.; Albrecht, D.; Cartron, M.; Hurd, A.; Foster, S.J. The effect of skin fatty acids on Staphylococcus aureus. Arch. Microbiol. 2015, 197, 245–267. [Google Scholar] [CrossRef] [Green Version]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef]
- Ouhara, K.; Komatsuzawa, H.; Kawai, T.; Nishi, H.; Fujiwara, T.; Fujiue, Y.; Kuwabara, M.; Sayama, K.; Hashimoto, K.; Sugai, M. Increased resistance to cationic antimicrobial peptide LL-37 in methicillin-resistant strains of Staphylococcus aureus. J. Antimicrob. Chemother. 2008, 61, 1266–1269. [Google Scholar] [CrossRef]
- Peschel, A.; Jack, R.W.; Otto, M.; Collins, L.V.; Staubitz, P.; Nicholson, G.; Kalbacher, H.; Nieuwenhuizen, W.F.; Jung, G.; Tarkowski, A.; et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J. Exp. Med. 2001, 193, 1067–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristian, S.A.; Durr, M.; van Strijp, J.A.; Neumeister, B.; Peschel, A. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect. Immun. 2003, 71, 546–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, L.; Alder, J.D.; Silverman, J.A. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2006, 50, 2137–2145. [Google Scholar] [CrossRef] [Green Version]
- Jones, T.; Yeaman, M.R.; Sakoulas, G.; Yang, S.J.; Proctor, R.A.; Sahl, H.G.; Schrenzel, J.; Xiong, Y.Q.; Bayer, A.S. Failures in clinical treatment of Staphylococcus aureus Infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob. Agents Chemother. 2008, 52, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falanga, A.; Nigro, E.; De Biasi, M.G.; Daniele, A.; Morelli, G.; Galdiero, S.; Scudiero, O. Cyclic Peptides as Novel Therapeutic Microbicides: Engineering of Human Defensin Mimetics. Molecules 2017, 20, 1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scudiero, O.; Nigro, E.; Cantisani, M.; Colavita, I.; Leone, M.; Mercurio, F.A.; Galdiero, M.; Pessi, A.; Daniele, A.; Salvatore, F.; et al. Design and activity of a cyclic mini-β-defensin analog: A novel antimicrobial tool. Int. J. Nanomed. 2015, 10, 6523–6539. [Google Scholar]
- Colavita, I.; Nigro, E.; Sarnataro, D.; Scudiero, O.; Granata, V.; Daniele, A.; Zagari, A.; Pessi, A.; Salvatore, F. Membrane protein 4F2/CD98 is a cell surface receptor involved in the internalization and trafficking of human β-Defensin 3 in epithelial cells. Chem. Biol. 2015, 2, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Pero, R.; Angrisano, T.; Brancaccio, M.; Falanga, A.; Lombardi, L.; Natale, F.; Laneri, S.; Lombardo, B.; Galdiero, S.; Scudiero, O. Beta-defensins and analogs in Helicobacter pylori infections: mRNA expression levels, DNA methylation, and antibacterialactivity. PLoS ONE 2019, 14, e0222295. [Google Scholar] [CrossRef] [Green Version]
- Pero, R.; Brancaccio, M.; Laneri, S.; De Biasi, M.G.; Lombardo, B.; Scudiero, O. A Novel View of Human Helicobacter pylori Infections: Interplay between Microbiota and Beta-Defensins. Biomolecules 2019, 9, E237. [Google Scholar] [CrossRef] [Green Version]
- Pero, R.; Coretti, L.; Nigro, E.; Lembo, F.; Laneri, S.; Lombardo, B.; Daniele, A.; Scudiero, O. β-Defensins in the Fight against Helicobacter pylori. Molecules 2017, 22, E424. [Google Scholar] [CrossRef] [PubMed]

| Defensin Type | Cellular Skin Production | Mechanism of S. aureus Evasion | References Doi Number |
|---|---|---|---|
| α-Defensins | Neutrophils | Staphylokinase, MprF, dltABCD operon | [73,74,75,76] |
| HBD2 | Keratinocytes, macrophages, and dendritic cells | IsdA, dltABCD | [77,78,79] |
| HBD3 | Keratinocytes | dltABCD operon | [79,80,81,82,83,84,85] |
| HBD4 | Keratinocytes | SAEC 6043 | [79] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scudiero, O.; Brancaccio, M.; Mennitti, C.; Laneri, S.; Lombardo, B.; De Biasi, M.G.; De Gregorio, E.; Pagliuca, C.; Colicchio, R.; Salvatore, P.; et al. Human Defensins: A Novel Approach in the Fight against Skin Colonizing Staphylococcus aureus. Antibiotics 2020, 9, 198. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9040198
Scudiero O, Brancaccio M, Mennitti C, Laneri S, Lombardo B, De Biasi MG, De Gregorio E, Pagliuca C, Colicchio R, Salvatore P, et al. Human Defensins: A Novel Approach in the Fight against Skin Colonizing Staphylococcus aureus. Antibiotics. 2020; 9(4):198. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9040198
Chicago/Turabian StyleScudiero, Olga, Mariarita Brancaccio, Cristina Mennitti, Sonia Laneri, Barbara Lombardo, Margherita G. De Biasi, Eliana De Gregorio, Chiara Pagliuca, Roberta Colicchio, Paola Salvatore, and et al. 2020. "Human Defensins: A Novel Approach in the Fight against Skin Colonizing Staphylococcus aureus" Antibiotics 9, no. 4: 198. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9040198





