Adapting a Phage to Combat Phage Resistance
Abstract
:1. Introduction
2. Results
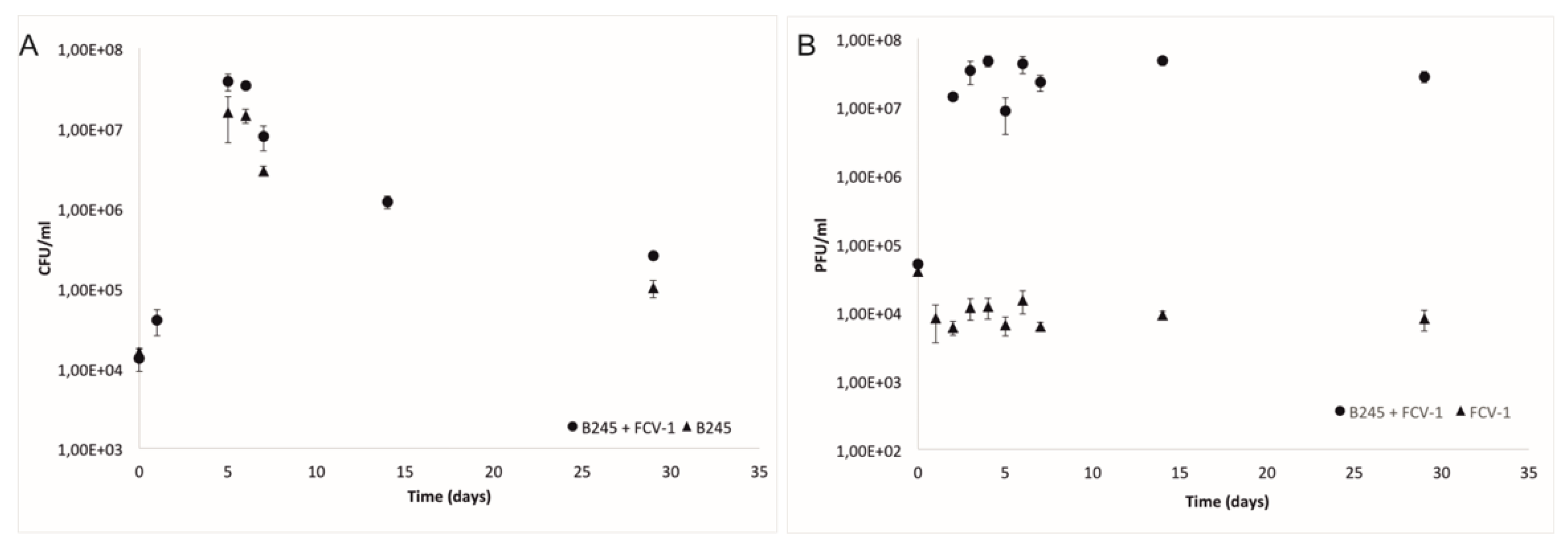
2.1. Bacteria and Phage during the Experiment
2.2. Phage Resistance Detected by Morphotype and CRISPR Spacer Acquisition
2.3. Phage Infectivity against the Evolved Bacteria
2.4. Ability to Infect Phage Resistant Clones Has a Direct Cost on Phage Adsorption
2.5. Nucleotide Level Differences Show Changes in Putative Tail Proteins
3. Discussion
4. Materials and Methods
4.1. Bacteria and Phage
4.2. Coevolution Experiment in Lake Water
4.3. Sampling
4.4. Infectivity of Isolated Phage
4.5. Adsorption Test
4.6. Sequencing of the Phage Genome
4.7. CRISPR Spacer Array Sequencing
4.8. Data Availability
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rostøl, J.T.; Marraffini, L. (ph)ighting phages: How bacteria resist their parasites. Cell Host Microbe. 2019, 25, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Hampton, H.G.; Watson, B.N.J.; Fineran, P.C. The arms race between bacteria and their phage foes. Nature 2020, 577, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Heierson, A.; Sidén, I.; Kivaisi, A.; Boman, H.G. Bacteriophage-resistant mutants of Bacillus thuringiensis with decreased virulence in pupae of Hyalophora cecropia. J. Bacteriol. 1986, 167, 18–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capparelli, R.; Nocerino, N.; Iannaccone, M.; Ercolini, D.; Parlato, M.; Chiara, M.; Iannelli, D. Bacteriophage therapy of Salmonella enterica: A fresh appraisal of bacteriophage therapy. J. Infect Dis. 2010, 201, 52–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friman, V.-P.; Hiltunen, T.; Jalasvuori, M.; Lindstedt, C.; Laanto, E.; Örmälä, A.-M.; Laakso, J.; Mappes, J.; Bamford, J.K. High temperature and bacteriophages can indirectly select for bacterial pathogenicity in environmental reservoirs. PLoS ONE 2011, 6, e17651. [Google Scholar] [CrossRef]
- Samson, J.E.; Magadán, A.H.; Sabri, M.; Moineau, S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013, 11, 675–687. [Google Scholar] [CrossRef]
- Hwang, S.; Maxwell, K.L. Meet the Anti-CRISPRs: Widespread Protein Inhibitors of CRISPR-Cas Systems. CRISPR J. 2019, 2, 23–30. [Google Scholar] [CrossRef]
- Rohde, C.; Resch, G.; Pirnay, J.-P.; Blasdel, B.G.; Debarbieux, L.; Gelman, D.; Górski, A.; Hazan, R.; Huys, I.; Kakabadze, E.; et al. Expert opinion on three phage therapy related topics: Bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 2018, 10, 178. [Google Scholar] [CrossRef] [Green Version]
- Betts, A.; Vasse, M.; Kaltz, O.; Hochberg, M.E. Back to the future: Evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evol. Appl. 2013, 6, 1054–1063. [Google Scholar]
- Friman, V.P.; Soanes-Brown, D.; Sierocinski, P.; Molin, S.; Johansen, H.K.; Merabishvili, M.; Pirnay, J.P.; De Vos, D.; Buckling, A. Pre-adapting parasitic phages to a pathogen leads to increased pathogen clearance and lowered resistance evolution with Pseudomonas aeruginosa cystic fibrosis bacterial isolates. J. Evol. Biol. 2016, 29, 188–198. [Google Scholar] [CrossRef] [Green Version]
- Declercq, A.M.; Haesebrouck, F.; Van den Broeck, W.; Bossier, P.; Decostere, A. Columnaris disease in fish: A review with emphasis on bacterium-host interactions. Vet. Res. 2013, 44, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laanto, E.; Bamford, J.K.H.; Laakso, J.; Sundberg, L.-R. Phage-driven loss of virulence in a fish pathogenic bacterium. PLoS ONE. 2012, 7, e53157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penttinen, R.; Hoikkala, V.; Sundberg, L.-R. Gliding Motility and Expression of Motility-Related Genes in Spreading and Non-Spreading Colonies of Flavobacterium columnare. Front. Microbiol. 2018, 9, 525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- León, M.; Bastías, R. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol. 2015, 6, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laanto, E.; Hoikkala, V.; Ravantti, J.; Sundberg, L.-R. Long-term genomic coevolution of host-parasite interaction in the natural environment. Nat. Commun. 2017, 8, 111. [Google Scholar] [CrossRef] [Green Version]
- Hoikkala, V.; Ravantti, J.J.; Diez-Villasenor, C.; Tiirola, M.; Conrad, R.; McBride, M.J.; Sundberg, L.R. Cooperation between CRISPR-Cas types enables adaptation in an RNA-targeting system. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Laanto, E.; Sundberg, L.-R.; Bamford, J.K.H. Phage specificity of the freshwater fish pathogen Flavobacterium columnare. Appl. Environ. Microbiol. 2011, 77, 7868–7872. [Google Scholar] [CrossRef] [Green Version]
- Sundberg, L.-R.; Kunttu, H.M.T.; Valtonen, E.T. Starvation can diversify the population structure and virulence strategies of an environmentally transmitting fish pathogen. BMC Microbiol. 2014, 14, 67. [Google Scholar] [CrossRef] [Green Version]
- Weitz, J.S.; Hartman, H.; Levin, S.A. Coevolutionary arms races between bacteria and bacteriophage. Proc. Natl. Acad. Sci. USA 2005, 102, 9535–9540. [Google Scholar] [CrossRef] [Green Version]
- Stern, A.; Sorek, R. The phage-host arms race: Shaping the evolution of microbes. Bioessays 2011, 33, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Górski, A.; Międzybrodzki, R.; Węgrzyn, G.; Jończyk-Matysiak, E.; Borysowski, J. Weber-Dąbrowska, B. Phage therapy: Current status and perspectives. Med. Res. Rev. 2020, 40, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Laanto, E.; Bamford, J.K.H.; Ravantti, J.J.; Sundberg, L.-R. The use of phage FCL-2 as an alternative to chemotherapy against columnaris disease in aquaculture. Front Microbiol. 2015, 6, 829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, G.M.F.; Laanto, E.; Ashrafi, R.; Sundberg, L.-R. Bacteriophage Adherence to Mucus Mediates Preventive Protection against Pathogenic Bacteria. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Pascua, L.d.C.; Buckling, A. Increasing productivity accelerates host-parasite coevolution. J. Evol. Biol. 2008, 21, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.; Brockhurst, M.A. Ecological and evolutionary benefits of temperate phage: What does or doesn’t kill you makes you stronger. Bioessays 2017, 39, 1700112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohannan, B.J.M.; Lenski, R.E. The Relative Importance of Competition and Predation Varies with Productivity in a Model Community. Am. Nat. 2000, 156, 329–340. [Google Scholar] [CrossRef]
- Harrison, E.; Laine, A.-L.; Hietala, M.; Brockhurst, M.A. Rapidly fluctuating environments constrain coevolutionary arms races by impeding selective sweeps. Proc. Biol. Sci. 2013, 280, 20130937. [Google Scholar] [CrossRef] [Green Version]
- Van Houte, S.; Ekroth, A.K.E.; Broniewski, J.M.; Chabas, H.; Ashby, B.; Bondy-Denomy, J.; Gandon, S.; Boots, M.; Paterson, S.; Buckling, A.; et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 2016, 532, 385–388. [Google Scholar] [CrossRef] [Green Version]
- Meeske, A.J.; Nakandakari-Higa, S.; Marraffini, L.A. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 2019, 570, 241–245. [Google Scholar] [CrossRef]
- Nelson, S.S.; Bollampalli, S.; McBride, M.J. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 2008, 190, 2851–2857. [Google Scholar] [CrossRef] [Green Version]
- Shrivastava, A.; Rhodes, R.G.; Pochiraju, S.; Nakane, D.; McBride, M.J. Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J. Bacteriol. 2012, 194, 3678–3688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunttu, H.M.T.; Sundberg, L.-R.; Pulkkinen, K.; Valtonen, E.T. Environment may be the source of Flavobacterium columnare outbreaks at fish farms. Environ. Microbiol. Rep. 2012, 4, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Laakso, J.; Mappes, J.; Laanto, E.; Ketola, T.; Bamford, J.K.H.; Kunttu, H.; Sundberg, L.R. Association of colony morphotypes with virulence, growth and resistance against protozoan predation in the fish pathogen Flavobacterium columnare. FEMS Microbiol. Ecol. 2014, 89, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; van Raaij, M. Bacteriophage T4 long tail fiber domains. Biophys. Rev. 2018, 10, 463–471. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.Z.; Fokine, A.; Mahalingam, M.; Zhang, Z.; Garcia-Doval, C.; van Raaij, M.J.; Rossmann, M.G.; Rao, V.B. Molecular anatomy of the receptor binding module of a bacteriophage long tail fiber. PLoS Pathog. 2019, 15, e1008193. [Google Scholar] [CrossRef] [Green Version]
- Tétart, F.; Repoila, F.; Monod, C.; Krisch, H.M. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J. Mol. Biol. 1996, 258, 726–731. [Google Scholar] [CrossRef]
- Sergueev, K.V.; Filippov, A.A.; Farlow, J.; Su, W.; Kvachadze, L.; Balarjishvili, N.; Kutateladze, M.; Nikolich, M.P. Correlation of Host Range Expansion of Therapeutic Bacteriophage Sb-1 with Allele State at a Hypervariable Repeat Locus. Appl. Environ. Microbiol. 2019, 85, e01209-19. [Google Scholar] [CrossRef] [Green Version]
- Gallet, R.; Lenormand, T.; Wang, I.-N. Phenotypic stochasticity protects lytic bacteriophage populations from extinction during the bacterial stationary phase. Evolution 2012, 66, 3485–3494. [Google Scholar] [CrossRef]
- Holtzman, T.; Globus, R.; Molshanski-Mor, S.; Ben-Shem, A.; Yosef, I.; Qimron, U. A continuous evolution system for contracting the host range of bacteriophage T7. Sci. Rep. 2020, 10, 307. [Google Scholar] [CrossRef]
- Dunne, M.; Rupf, B.; Tala, M.; Qabrati, X.; Ernst, P.; Shen, Y.; Sumrall, E.; Heeb, L.; Plückthun, A.; Loessner, M.J.; et al. Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Rep. 2019, 29, 1336–1350.e4. [Google Scholar] [CrossRef] [Green Version]
- Kunttu, H.M.T.; Suomalainen, L.-R.; Jokinen, E.I.; Valtonen, E.T. Flavobacterium columnare colony types: Connection to adhesion and virulence? Microb. Pathog. 2009, 46, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.L.; Fryer, J.L.; Rohovec, J.S. Comparison of six media for the cultivation of Flexibacter columnaris. Fish Pathol. 1988, 23, 91–94. [Google Scholar] [CrossRef] [Green Version]
- Kropinski, A.M. Measurement of the rate of attachment of bacteriophage to cells. Methods Mol. Biol. 2009, 501, 151–155. [Google Scholar] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Phage Titer (pfu/mL) | ||
|---|---|---|
| Bacteria | FCV-1 | FCV-1.01 |
| C4 | 1,1 × 1010 | 1,1 × 1010 |
| 1 ancestral B245 | 1,3 × 1010 | 1,0 × 1010 |
| 2 (1 week), sensitive1 rhizoid | 6,0 × 107 | 1,4 × 1010 |
| 3 (1 week) resistant1 rhizoid | * | 1,1 × 1010 |
| 4 (1 week), resistant1 rough | NA | 1,0 × 1010 |
| 5 (1 week), resistant1 rhizoid | ** | 1,1 × 1010 |
| 6 (1 week), resistant1 rough | NA | 4,9 × 1009 |
| 7 (2 weeks), resistant1 rhizoid, spacer added | 0 | 2,0 × 1010 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laanto, E.; Mäkelä, K.; Hoikkala, V.; Ravantti, J.J.; Sundberg, L.-R. Adapting a Phage to Combat Phage Resistance. Antibiotics 2020, 9, 291. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9060291
Laanto E, Mäkelä K, Hoikkala V, Ravantti JJ, Sundberg L-R. Adapting a Phage to Combat Phage Resistance. Antibiotics. 2020; 9(6):291. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9060291
Chicago/Turabian StyleLaanto, Elina, Kati Mäkelä, Ville Hoikkala, Janne J. Ravantti, and Lotta-Riina Sundberg. 2020. "Adapting a Phage to Combat Phage Resistance" Antibiotics 9, no. 6: 291. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9060291





