The pathogen Mycoplasma dispar Shows High Minimum Inhibitory Concentrations for Antimicrobials Commonly Used for Bovine Respiratory Disease
Abstract
:1. Introduction
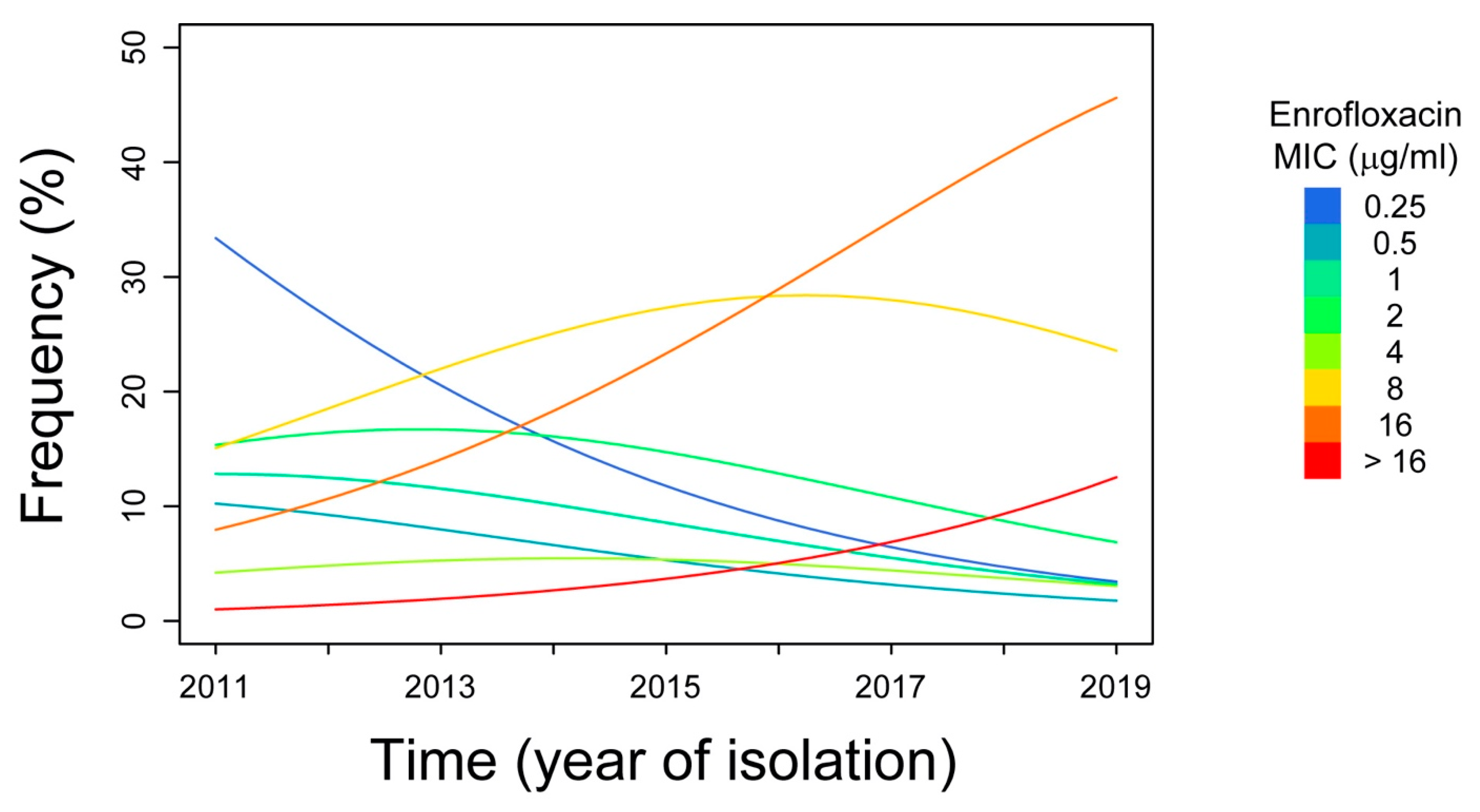
2. Results
3. Discussion
4. Material and Methods:
4.1. Mycoplasma Dispar Isolates Collection
4.2. M. dispar Isolates In Vitro Cultivation and Identification.
4.3. MIC Test for Mycoplasma dispar
4.4. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Gourlay, R.N.; Leach, R.H. A new mycoplasma species isolated from pneumonic lungs of calves (Mycoplasma dispar sp. nov.). J. Med. Microbiol. 1970, 3, 111–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- França Dias de Oliveira, B.A.; Gaeta, N.C.; Ribeiro, B.L.M.; Alemán, M.A.R.; Marques, L.M.; Timenetsky, J.; Melville, P.A.; Marques, J.A.; Marvulle, V.; Gregory, L. Determination of bacterial aetiologic factor on tracheobronchial lavage in relation to clinical signs of bovine respiratory disease. J. Med. Microbiol. 2016, 65, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Ter Laak, E.A.; Noordergraaf, J.H.; Dieltjes, R.P.J.W. Prevalence of Mycoplasmas in the Respiratory Tracts of Pneumonic Calves. J. Vet. Med. Ser. B 1992, 39, 553–562. [Google Scholar] [CrossRef]
- Laak, E.A.; Noordergraaf, J.H.; Boomsluiter, E. The Nasal Mycoplasmal Flora of Healthy Calves and Cows. J. Vet. Med. Ser. B 1992, 39, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, R.A.J.; Ayling, R.D.; McAuliffe, L. Bovine Respiratory Disease. In Mycoplasma Diseases of Ruminants; CABI: Wallingford, UK, 2008; pp. 132–168. ISBN 978-0-85199-012-5. [Google Scholar]
- Brown, D.R. Phylum XVI. Tenericutes Murray 1984a, 356VP. In Bergey’s Manual of Systematic Bacteriology; Noel, K.R., James, S.T., Brown, D.R., Brian, H.P., Bruce, P.J., Naomi, W.L., Wolfgang, L., Whitman, W.B., Eds.; Springer Science + Business Media, LLC: New York, NY, USA, 2010; Volume 4, pp. 567–639. [Google Scholar]
- Urban-Chmiel, R.; Grooms, D.L. Prevention and Control of Bovine Respiratory Disease. J. Livestock Sci. 2012, 3, 27–36. [Google Scholar]
- Gourlay, R.N.; Howard, C.J.; Thomas, L.H.; Wyld, S.G. Pathogenicity of some Mycoplasma and Acholeplasma species in the lungs of gnotobiotic calves. Res. Vet. Sci. 1979, 27, 233–237. [Google Scholar] [CrossRef]
- Friis, N.F. Mycoplasma dispar as a causative agent in pneumonia of calves. Acta Vet. Scand. 1980, 21, 34–42. [Google Scholar]
- Virtala, A.M.K.; Mechor, G.D.; Gröhn, Y.T.; Erb, H.N.; Dubovi, E.J. Epidemiologic and pathologic characteristics of respiratory tract disease in dairy heifers during the first three months of life. J. Am. Vet. Med. Assoc. 1996, 208, 2035–2042. [Google Scholar]
- Van Donkersgoed, J.; Ribble, C.S.; Boyer, L.G.; Townsend, H.G. Epidemiological study of enzootic pneumonia in dairy calves in Saskatchewan. Can. J. Vet. Res. 1993, 57, 247–254. [Google Scholar]
- Chen, S.; Hao, H.; Yan, X.; Liu, Y.; Chu, Y. Genome-Wide Analysis of Mycoplasma dispar provides insights into putative virulence factors and phylogenetic relationships. G3 Genes Genomes Genet. 2019, 9, 317–325. [Google Scholar] [CrossRef] [Green Version]
- Thomas, L.H.; Howard, C.J.; Parsons, K.R.; Anger, H.S. Growth of Mycoplasma bovis in organ cultures of bovine foetal trachea and comparison with Mycoplasma dispar. Vet. Microbiol. 1987, 13, 189–200. [Google Scholar] [CrossRef]
- Tajima, M.; Yagihashi, T. Interaction of Mycoplasma hyopneumoniae with the porcine respiratory epithelium as observed by electron microscopy. Infect. Immun. 1982, 37, 1162–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, R.A.; Rosenbusch, R.F. Impaired Tracheobronchial Clearance of Bacteria in Calves Infected with Mycoplasma dispar. J. Vet. Med. Ser. B 1994, 41, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.A.; Wannemuehler, M.J.; Rosenbusch, R.F. Interaction of Mycoplasma dispar with bovine alveolar macrophages. Infect. Immun. 1992, 60, 2914–2919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Casal, J. Pathogenesis and Virulence of Mycoplasma bovis. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 269–278. [Google Scholar] [CrossRef]
- Dedonder, K.D.; Apley, M.D. A literature review of antimicrobial resistance in Pathogens associated with bovine respiratory disease. Anim. Heal. Res. Rev. 2015, 16, 125–134. [Google Scholar] [CrossRef]
- Anholt, R.M.; Klima, C.; Allan, N.; Matheson-Bird, H.; Schatz, C.; Ajitkumar, P.; Otto, S.J.; Peters, D.; Schmid, K.; Olson, M.; et al. Antimicrobial Susceptibility of Bacteria That Cause Bovine Respiratory Disease Complex in Alberta, Canada. Front. Vet. Sci. 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- Ter Laak, E.A.; Noordergraaf, J.H.; Verschure, M.H. Susceptibilities of Mycoplasma bovis, Mycoplasma dispar, and Ureaplasma diversum strains to antimicrobial agents in vitro. Antimicrob. Agents Chemother. 1993, 37, 317–321. [Google Scholar] [CrossRef] [Green Version]
- Ayling, R.D.; Rosales, R.S.; Barden, G.; Gosney, F.L. Changes in antimicrobial susceptibility of Mycoplasma bovis isolates from Great Britain. Vet. Rec. 2014, 175, 486. [Google Scholar] [CrossRef]
- Gautier-Bouchardon, A.V. Antimicrobial Resistance in Mycoplasma spp. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Klein, U.; de Jong, A.; Youala, M.; El Garch, F.; Stevenin, C.; Moyaert, H.; Rose, M.; Catania, S.; Gyuranecz, M.; Pridmore, A.; et al. New antimicrobial susceptibility data from monitoring of Mycoplasma bovis isolated in Europe. Vet. Microbiol. 2019, 238. [Google Scholar] [CrossRef] [PubMed]
- Gautier-Bouchardon, A.V.; Ferré, S.; Le Grand, D.; Paoli, A.; Gay, E.; Poumarat, F. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portis, E.; Lindeman, C.; Johansen, L.; Stoltman, G. A ten-year (2000–2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex-Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni-in the United States and Canada. J. Vet. Diagnostic Investig. 2012, 24, 932–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannan, P.C.T. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. Vet. Res. 2000, 31, 373–395. [Google Scholar] [CrossRef] [Green Version]
- Catania, S.; Bottinelli, M.; Fincato, A.; Gastaldelli, M.; Barberio, A.; Gobbo, F.; Vicenzoni, G. Evaluation of Minimum Inhibitory Concentrations for 154 Mycoplasma synoviae isolates from Italy collected during 2012–2017. PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [Green Version]
- Lysnyansky, I.; Gerchman, I.; Mikula, I.; Gobbo, F.; Catania, S.; Levisohn, S. Molecular characterization of acquired enrofloxacin resistance in Mycoplasma synoviae field isolates. Antimicrob. Agents Chemother. 2013, 57, 3072–3077. [Google Scholar] [CrossRef] [Green Version]
- Sirand-Pugnet, P.; Citti, C.; Barré, A.; Blanchard, A. Evolution of mollicutes: Down a bumpy road with twists and turns. Res. Microbiol. 2007, 158, 754–766. [Google Scholar] [CrossRef]
- Vasconcelos, A.T.R.; Ferreira, H.B.; Bizarro, C.V.; Bonatto, S.L.; Carvalho, M.O.; Pinto, P.M.; Almeida, D.F.; Almeida, L.G.P.; Almeida, R.; Alves-Filho, L.; et al. Swine and poultry pathogens: The complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 2005, 187, 5568–5577. [Google Scholar] [CrossRef] [Green Version]
- Faucher, M.; Nouvel, L.X.; Dordet-Frisoni, E.; Sagné, E.; Baranowski, E.; Hygonenq, M.C.; Marenda, M.S.; Tardy, F.; Citti, C. Mycoplasmas under experimental antimicrobial selection: The unpredicted contribution of horizontal chromosomal transfer. PLoS Genet. 2019, 15. [Google Scholar] [CrossRef] [Green Version]
- Hannan, P.C.T.; O’Hanlon, P.J.; Rogers, N.H. In vitro evaluation of various quinolone antibacterial agents against veterinary mycoplasmas and porcine respiratory bacterial pathogens. Res. Vet. Sci. 1989, 46, 202–211. [Google Scholar] [CrossRef]
- Barberio, A.; Flaminio, B.; De Vliegher, S.; Supré, K.; Kromker, V.; Garbarino, C.; Arrigoni, N.; Zanardi, G.; Bertocchi, L.; Gobbo, F.; et al. Short communication: In vitro antimicrobial susceptibility of Mycoplasma bovis isolates identified in milk from dairy cattle in Belgium, Germany, and Italy. J. Dairy Sci. 2016, 99, 6578–6584. [Google Scholar] [CrossRef] [PubMed]
- Rosenbusch, R.F.; Kinyon, J.M.; Apley, M.; Funk, N.D.; Smith, S.; Hoffman, L.J. In vitro antimicrobial inhibition profiles of Mycoplasma bovis isolates recovered from various regions of the United States from 2002 to 2003. J. Vet. Diagnostic Investig. 2005, 17, 436–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francoz, D.; Fortin, M.; Fecteau, G.; Messier, S. Determination of Mycoplasma bovis susceptibilities against six antimicrobial agents using the E test method. Vet. Microbiol. 2005, 105, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Devriese, L.A.; Haesebrouck, F. Antibiotic Susceptibility Testing of Mycoplasma bovis using Tween 80 Hydrolysis as an Indicator of Growth. J. Vet. Med. Ser. B 1991, 38, 781–783. [Google Scholar] [CrossRef]
- Friis, N.F.; Szancer, J. Sensitivity of certain porcine and bovine mycoplasmas to antimicrobial agents in a liquid medium test compared to a disc assay. Acta Vet. Scand. 1994, 35, 389–394. [Google Scholar]
- Bekő, K.; Felde, O.; Sulyok, K.M.; Kreizinger, Z.; Hrivnák, V.; Kiss, K.; Biksi, I.; Jerzsele, Á.; Gyuranecz, M. Antibiotic susceptibility profiles of Mycoplasma hyorhinis strains isolated from swine in Hungary. Vet. Microbiol. 2019, 228, 196–201. [Google Scholar] [CrossRef]
- Klein, U.; de Jong, A.; Moyaert, H.; El Garch, F.; Leon, R.; Richard-Mazet, A.; Rose, M.; Maes, D.; Pridmore, A.; Thomson, J.R.; et al. Antimicrobial susceptibility monitoring of Mycoplasma hyopneumoniae and Mycoplasma bovis isolated in Europe. Vet. Microbiol. 2017, 204, 188–193. [Google Scholar] [CrossRef]
- Gigueré, S. Lincosamides, pleuromutilins and streptogramins. In Antimicrobial Therapy in Veterinary Medicine; Gigueré, S., Prescott, J.F., Dowling, P.M., Eds.; John Wiley & Sons, Inc.: Ames, IA, USA, 2013; pp. 199–210. [Google Scholar]
- McAuliffe, L.; Ellis, R.J.; Lawes, J.R.; Ayling, R.D.; Nicholas, R.A.J. 16S rDNA PCR and denaturing gradient gel electrophoresis; a single generic test for detecting and differentiating Mycoplasma species. J. Med. Microbiol. 2005, 54, 731–739. [Google Scholar] [CrossRef] [Green Version]
- Clinical and Laboratory Standards Institute. M43-A Methods for Antimicrobial Susceptibility Testing for Human Mycoplasmas; Approved Guideline; Clinical and Laboratory Standards Institute: Annapolis Junction, MD, USA, 2011; ISBN 1562387693. [Google Scholar]
- Blodgett, R. FDA’s Bacteriological Analytical Manual, Appendix 2: Most Probable Number from Serial Dilutions; US Food and Drug Administration: Silver Spring, MD, USA, 2010.
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019.
- Hothorn, T.; Hornik, K.; van de Wiel, M.A.; Zeileis, A. Implementing a class of permutation tests: The coin package. J Stat Softw. 2008, 28, 1–23. [Google Scholar] [CrossRef]
- Christensen, R.H.B. Ordinal—Regression Models for Ordinal Data. R Package Version 2019.12-10. 2019. Available online: https://CRAN.R-project.org/package=ordinal (accessed on 15 December 2019).
| Antimicrobial | MIC Values (µg/mL) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.0078125 | 0.015625 | 0.03125 | 0.0625 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | |
| Enrofloxacin | 5 | 2 | 3 | 5 | 2 | 11 50 | 11 90 | 2 * | ||||||
| Oxytetracycline | 10 | 6 | 5 50 | 5 | 3 | 5 | 1 90 | 6 | ||||||
| Doxycycline | 7 | 4 | 4 | 18 50 | 3 | 2 90 | 2 | 1 | ||||||
| Erythromycin | 1 | 1 | 39 50-90 | |||||||||||
| Tylosin | 2 | 3 | 4 | 16 50 | 5 | 3 | 3 | 5 90 | ||||||
| Tilmicosin | 1 | 2 | 2 | 6 | 7 | 12 50 | 11 90 | |||||||
| Spiramycin | 12 | 8 | 11 50 | 2 | 1 | 7 90 | ||||||||
| Tiamulin | 1 | 1 | 2 | 5 | 19 50 | 7 | 6 90 | |||||||
| Florfenicol | 32 50 | 9 90 | ||||||||||||
| Lincomycin | 17 | 14 50 | 2 | 1 | 7 90 | |||||||||
| Antibiotic | Z-Value | p-Value |
|---|---|---|
| Doxicycline | +0.79 | 0.428 |
| Enrofloxacin | +2.39 | 0.0168 |
| Erythromycin | −1.68 | 0.0925 |
| Florfenicol | +0.14 | 0.886 |
| Lyncomycin | -1.68 | 0.0928 |
| Oxytetracycline | +0.39 | 0.695 |
| Spiramycin | −1.84 | 0.0654 |
| Tiamulin | −2.06 | 0.0389 |
| Tylosin | −2.21 | 0.0273 |
| Tilmicosin | −1.43 | 0.152 |
| Tylvalosin | +0.56 | 0.574 |
| Coefficients | Estimate | Std. Error | Z-Value | p-Value |
|---|---|---|---|---|
| Time | 0.331 | 0.136 | 2.44 | 0.0147 |
| Threshold coefficients | ||||
| 0.25|0.5 | −0.691 | 0.689 | −3.056 | |
| 0.5|1 | −0.257 | 0.663 | −0.388 | |
| 1|2 | 0.256 | 0.662 | 0.391 | |
| 2|4 | 0.933 | 0.683 | 1.367 | |
| 4|8 | 1.151 | 0.686 | 1.677 | |
| 8|16 | 2.319 | 0.725 | 3.198 | |
| 16|>16 | 4.591 | 0.992 | 4.629 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bottinelli, M.; Merenda, M.; Gastaldelli, M.; Picchi, M.; Stefani, E.; Nicholas, R.A.J.; Catania, S. The pathogen Mycoplasma dispar Shows High Minimum Inhibitory Concentrations for Antimicrobials Commonly Used for Bovine Respiratory Disease. Antibiotics 2020, 9, 460. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9080460
Bottinelli M, Merenda M, Gastaldelli M, Picchi M, Stefani E, Nicholas RAJ, Catania S. The pathogen Mycoplasma dispar Shows High Minimum Inhibitory Concentrations for Antimicrobials Commonly Used for Bovine Respiratory Disease. Antibiotics. 2020; 9(8):460. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9080460
Chicago/Turabian StyleBottinelli, Marco, Marianna Merenda, Michele Gastaldelli, Micaela Picchi, Elisabetta Stefani, Robin A. J. Nicholas, and Salvatore Catania. 2020. "The pathogen Mycoplasma dispar Shows High Minimum Inhibitory Concentrations for Antimicrobials Commonly Used for Bovine Respiratory Disease" Antibiotics 9, no. 8: 460. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics9080460






