Regulated DNA Methylation and the Circadian Clock: Implications in Cancer
Abstract
:1. DNA Methylation
2. Genome-Wide Analyses
3. The Circadian Clock
4. The Clock and Chromatin
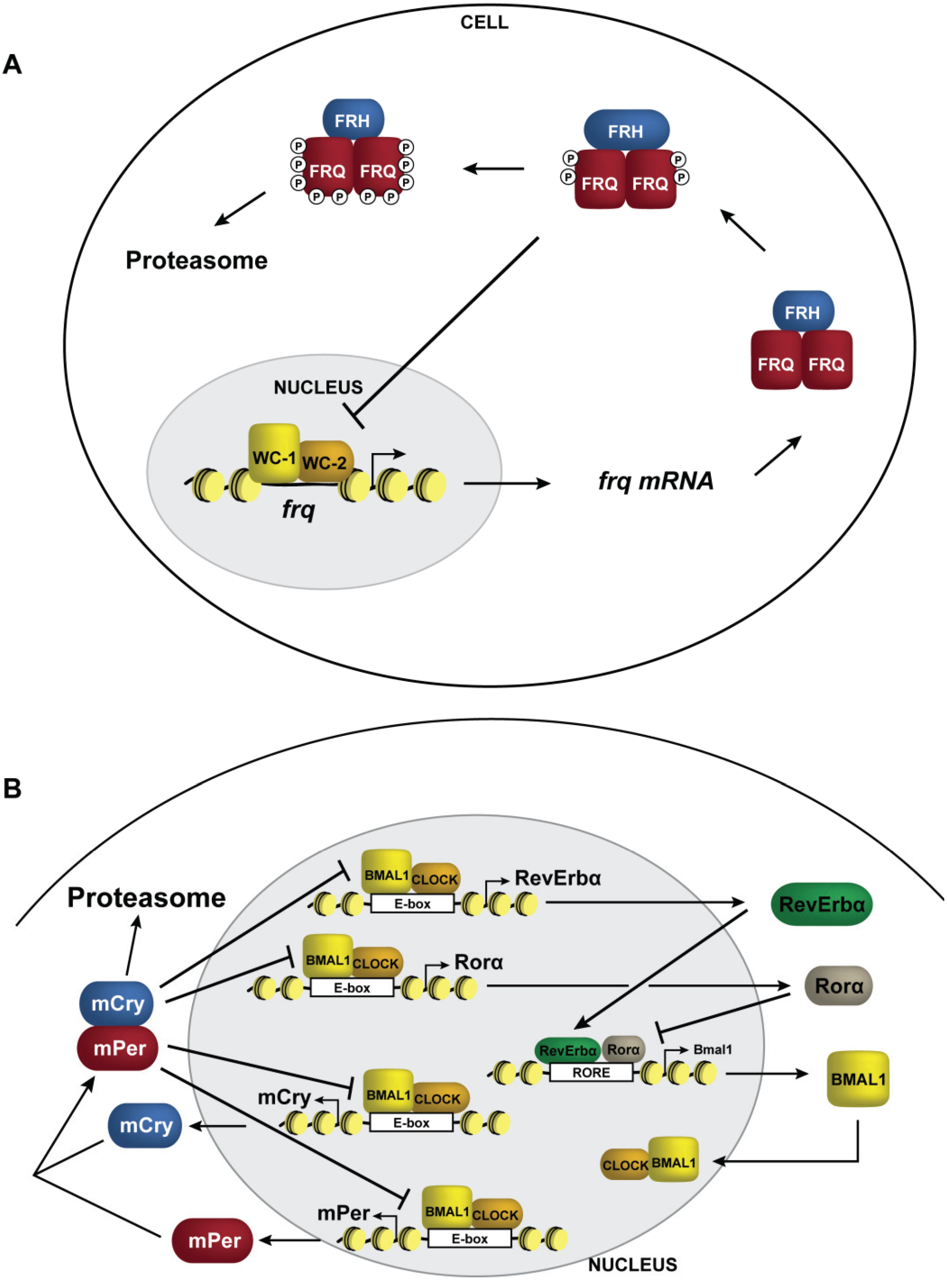
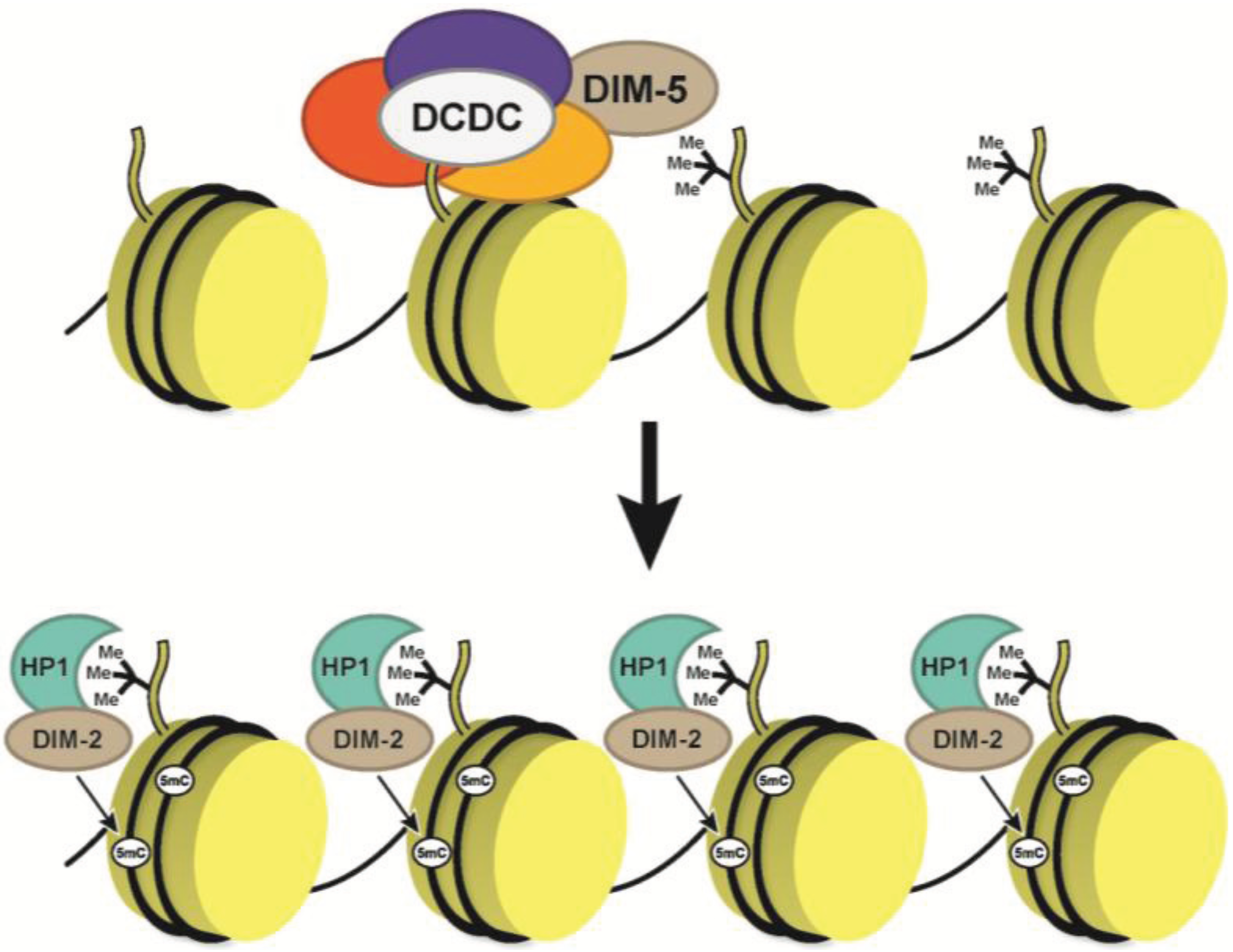
5. DNA Methylation in Neurospora
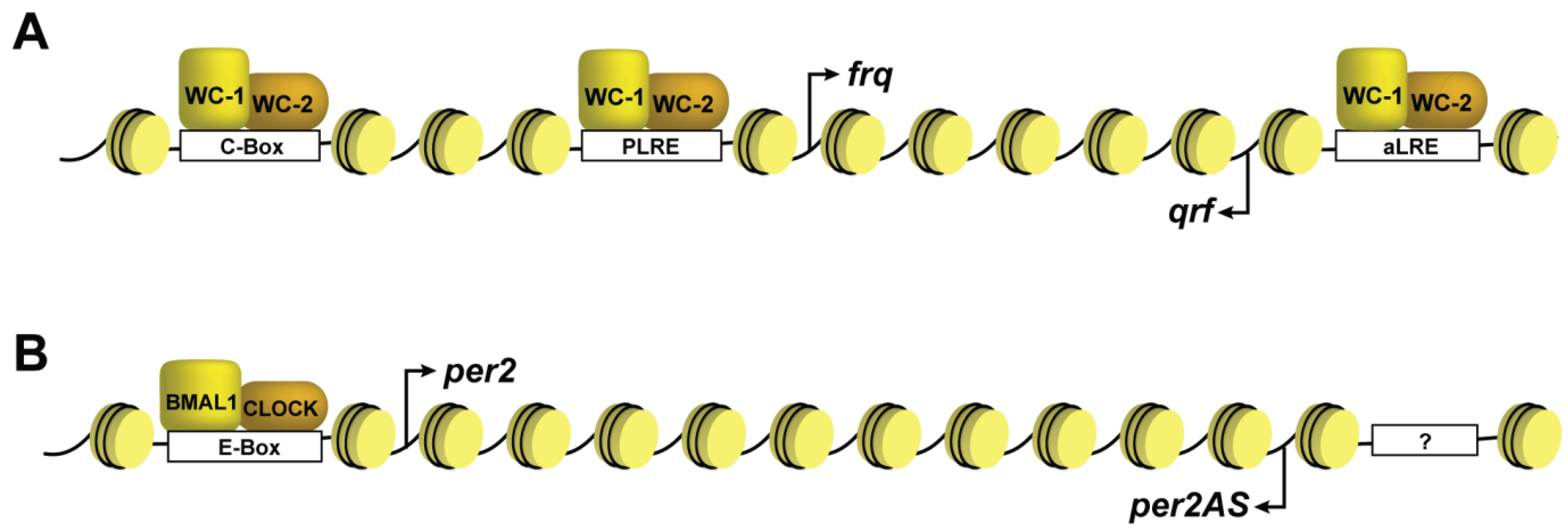
6. Dynamic DNA Methylation at Circadian Clock Genes

7. DNA Methylation, the Circadian Clock and Cancer
8. Misregulated Genes and DNA Methylation: The Chicken or the Egg?
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bestor, T.H. Cloning of a mammalian DNA methyltransferase. Gene 1988, 74, 9–12. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Meissner, A.; Lander, E.S. The mammalian epigenome. Cell 2007, 128, 669–681. [Google Scholar] [CrossRef]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.K.; O’Donnell, A.H.; Bestor, T.H. Mammalian cytosine methylation at a glance. J. Cell Sci. 2009, 122, 2787–2791. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Bestor, T.H. Unanswered questions about the role of promoter methylation in carcinogenesis. Ann. N. Y. Acad. Sci. 2003, 983, 22–27. [Google Scholar] [CrossRef]
- Ehrlich, M.; Lacey, M. DNA hypomethylation and hemimethylation in cancer. Adv. Exp. Med. Biol. 2013, 754, 31–56. [Google Scholar]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Mathieu, O.; Bender, J. RNA-directed DNA methylation. J. Cell Sci. 2004, 117, 4881–4888. [Google Scholar] [CrossRef]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef]
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992, 69, 915–926. [Google Scholar] [CrossRef]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar]
- Capuano, F.; Mulleder, M.; Kok, R.; Blom, H.J.; Ralser, M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in saccharomyces cerevisiae, schizosaccharomyces pombe, and other yeast species. Anal. Chem. 2014, 86, 3697–3702. [Google Scholar] [CrossRef]
- Walsh, C.P.; Bestor, T.H. Cytosine methylation and mammalian development. Genes Dev. 1999, 13, 26–34. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef]
- Belden, W.J.; Lewis, Z.A.; Selker, E.U.; Loros, J.J.; Dunlap, J.C. CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet. 2011, 7, e1002166. [Google Scholar] [CrossRef]
- Azzi, A.; Dallmann, R.; Casserly, A.; Rehrauer, H.; Patrignani, A.; Maier, B.; Kramer, A.; Brown, S.A. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat. Neurosci. 2014, 17, 377–382. [Google Scholar] [CrossRef]
- Shen, H.; He, H.; Li, J.; Chen, W.; Wang, X.; Guo, L.; Peng, Z.; He, G.; Zhong, S.; Qi, Y.; et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 2012, 24, 875–892. [Google Scholar] [CrossRef]
- Bell-Pedersen, D.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005, 6, 544–556. [Google Scholar]
- Hardin, P.E.; Panda, S. Circadian timekeeping and output mechanisms in animals. Curr. Opin. Neurobiol. 2013, 23, 724–731. [Google Scholar] [CrossRef]
- Schibler, U.; Sassone-Corsi, P. A web of circadian pacemakers. Cell 2002, 111, 919–922. [Google Scholar] [CrossRef]
- Dunlap, J.C. Molecular bases for circadian clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef]
- Ko, C.H.; Takahashi, J.S. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006, 15, R271–R277. [Google Scholar] [CrossRef]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef]
- Bass, J.; Takahashi, J.S. Circadian integration of metabolism and energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef]
- Loros, J.J.; Dunlap, J.C.; Larrondo, L.F.; Shi, M.; Belden, W.J.; Gooch, V.D.; Chen, C.H.; Baker, C.L.; Mehra, A.; Colot, H.V.; et al. Circadian output, input, and intracellular oscillators: Insights into the circadian systems of single cells. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 201–214. [Google Scholar] [CrossRef]
- Sahar, S.; Sassone-Corsi, P. Metabolism and cancer: The circadian clock connection. Nat. Rev. Cancer 2009, 9, 886–896. [Google Scholar] [CrossRef]
- McClung, C.R. The genetics of plant clocks. Adv. Genet. 2011, 74, 105–139. [Google Scholar]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef]
- Young, M.W. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu. Rev. Biochem. 1998, 67, 135–152. [Google Scholar] [CrossRef]
- Panda, S.; Antoch, M.P.; Miller, B.H.; Su, A.I.; Schook, A.B.; Straume, M.; Schultz, P.G.; Kay, S.A.; Takahashi, J.S.; Hogenesch, J.B. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002, 109, 307–320. [Google Scholar] [CrossRef]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef]
- Storch, K.F.; Lipan, O.; Leykin, I.; Viswanathan, N.; Davis, F.C.; Wong, W.H.; Weitz, C.J. Extensive and divergent circadian gene expression in liver and heart. Nature 2002, 417, 78–83. [Google Scholar] [CrossRef]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef]
- Hughes, M.E.; DiTacchio, L.; Hayes, K.R.; Vollmers, C.; Pulivarthy, S.; Baggs, J.E.; Panda, S.; Hogenesch, J.B. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009, 5, e1000442. [Google Scholar] [CrossRef]
- Brunner, M.; Kaldi, K. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol. Microbiol. 2008, 68, 255–262. [Google Scholar] [CrossRef]
- Liu, Y.; Bell-Pedersen, D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot. Cell 2006, 5, 1184–1193. [Google Scholar] [CrossRef]
- Crosthwaite, S.K.; Dunlap, J.C.; Loros, J.J. Neurospora wc-1 and wc-2: Transcription, photoresponses, and the origins of circadian rhythmicity. Science 1997, 276, 763–769. [Google Scholar] [CrossRef]
- Bunger, M.K.; Wilsbacher, L.D.; Moran, S.M.; Clendenin, C.; Radcliffe, L.A.; Hogenesch, J.B.; Simon, M.C.; Takahashi, J.S.; Bradfield, C.A. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 2000, 103, 1009–1017. [Google Scholar] [CrossRef]
- Hogenesch, J.B.; Gu, Y.Z.; Jain, S.; Bradfield, C.A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 1998, 95, 5474–5479. [Google Scholar] [CrossRef]
- Antoch, M.P.; Song, E.J.; Chang, A.M.; Vitaterna, M.H.; Zhao, Y.; Wilsbacher, L.D.; Sangoram, A.M.; King, D.P.; Pinto, L.H.; Takahashi, J.S. Functional identification of the mouse circadian clock gene by transgenic BAC rescue. Cell 1997, 89, 655–667. [Google Scholar] [CrossRef]
- Cheng, P.; He, Q.; Wang, L.; Liu, Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 2005, 19, 234–241. [Google Scholar] [CrossRef]
- Aronson, B.D.; Johnson, K.A.; Dunlap, J.C. Circadian clock locus frequency: Protein encoded by a single open reading frame defines period length and temperature compensation. Proc. Natl. Acad. Sci. USA 1994, 91, 7683–7687. [Google Scholar] [CrossRef]
- Sato, T.K.; Yamada, R.G.; Ukai, H.; Baggs, J.E.; Miraglia, L.J.; Kobayashi, T.J.; Welsh, D.K.; Kay, S.A.; Ueda, H.R.; Hogenesch, J.B. Feedback repression is required for mammalian circadian clock function. Nat. Genet. 2006, 38, 312–319. [Google Scholar] [CrossRef]
- Zylka, M.J.; Shearman, L.P.; Weaver, D.R.; Reppert, S.M. Three period homologs in mammals: Differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 1998, 20, 1103–1110. [Google Scholar] [CrossRef]
- Darlington, T.K.; Wager-Smith, K.; Ceriani, M.F.; Staknis, D.; Gekakis, N.; Steeves, T.D.; Weitz, C.J.; Takahashi, J.S.; Kay, S.A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and time. Science 1998, 280, 1599–1603. [Google Scholar] [CrossRef]
- Tei, H.; Okamura, H.; Shigeyoshi, Y.; Fukuhara, C.; Ozawa, R.; Hirose, M.; Sakaki, Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 1997, 389, 512–516. [Google Scholar] [CrossRef]
- Kume, K.; Zylka, M.J.; Sriram, S.; Shearman, L.P.; Weaver, D.R.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M. MCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999, 98, 193–205. [Google Scholar] [CrossRef]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Baker, C.L.; Kettenbach, A.N.; Loros, J.J.; Gerber, S.A.; Dunlap, J.C. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol. Cell 2009, 34, 354–363. [Google Scholar] [CrossRef]
- Cha, J.; Yuan, H.; Liu, Y. Regulation of the activity and cellular localization of the circadian clock protein FRQ. J. Biol. Chem. 2011, 286, 11469–11478. [Google Scholar] [CrossRef]
- Diernfellner, A.C.; Querfurth, C.; Salazar, C.; Hofer, T.; Brunner, M. Phosphorylation modulates rapid nucleocytoplasmic shuttling and cytoplasmic accumulation of Neurospora clock protein FRQ on a circadian time scale. Genes Dev. 2009, 23, 2192–2200. [Google Scholar] [CrossRef]
- Nawathean, P.; Stoleru, D.; Rosbash, M. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol. Cell. Biol. 2007, 27, 5002–5013. [Google Scholar] [CrossRef]
- Ko, H.W.; Kim, E.Y.; Chiu, J.; Vanselow, J.T.; Kramer, A.; Edery, I. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3beta/SGG in circadian clocks. J. Neurosci. 2010, 30, 12664–12675. [Google Scholar] [CrossRef]
- St John, P.C.; Hirota, T.; Kay, S.A.; Doyle, F.J., 3rd. Spatiotemporal separation of PER and CRY posttranslational regulation in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 2040–2045. [Google Scholar]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Vollmers, C.; Schmitz, R.J.; Nathanson, J.; Yeo, G.; Ecker, J.R.; Panda, S. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 2012, 16, 833–845. [Google Scholar] [CrossRef]
- Aguilar-Arnal, L.; Hakim, O.; Patel, V.R.; Baldi, P.; Hager, G.L.; Sassone-Corsi, P. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat. Struct. Mol. Biol. 2013, 20, 1206–1213. [Google Scholar]
- Brown, S.A.; Ripperger, J.; Kadener, S.; Fleury-Olela, F.; Vilbois, F.; Rosbash, M.; Schibler, U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 2005, 308, 693–696. [Google Scholar] [CrossRef]
- Duong, H.A.; Robles, M.S.; Knutti, D.; Weitz, C.J. A molecular mechanism for circadian clock negative feedback. Science 2011, 332, 1436–1439. [Google Scholar] [CrossRef]
- Duong, H.A.; Weitz, C.J. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat. Struct. Mol. Biol. 2014, 21, 126–132. [Google Scholar] [CrossRef]
- Masri, S.; Zocchi, L.; Katada, S.; Mora, E.; Sassone-Corsi, P. The circadian clock transcriptional complex: Metabolic feedback intersects with epigenetic control. Ann. N. Y. Acad. Sci. 2012, 1264, 103–109. [Google Scholar] [CrossRef]
- Ramsey, K.M.; Yoshino, J.; Brace, C.S.; Abrassart, D.; Kobayashi, Y.; Marcheva, B.; Hong, H.K.; Chong, J.L.; Buhr, E.D.; Lee, C.; et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009, 324, 651–654. [Google Scholar] [CrossRef]
- Nakahata, Y.; Sahar, S.; Astarita, G.; Kaluzova, M.; Sassone-Corsi, P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009, 324, 654–657. [Google Scholar] [CrossRef]
- Kim, J.S.; Coon, S.L.; Blackshaw, S.; Cepko, C.L.; Moller, M.; Mukda, S.; Zhao, W.Q.; Charlton, C.G.; Klein, D.C. Methionine adenosyltransferase:adrenergic-cAMP mechanism regulates a daily rhythm in pineal expression. J. Biol. Chem. 2005, 280, 677–684. [Google Scholar] [CrossRef]
- Crosio, C.; Cermakian, N.; Allis, C.D.; Sassone-Corsi, P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 2000, 3, 1241–1247. [Google Scholar] [CrossRef]
- Etchegaray, J.P.; Lee, C.; Wade, P.A.; Reppert, S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 2003, 421, 177–182. [Google Scholar] [CrossRef]
- Curtis, A.M.; Seo, S.B.; Westgate, E.J.; Rudic, R.D.; Smyth, E.M.; Chakravarti, D.; FitzGerald, G.A.; McNamara, P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 2004, 279, 7091–7097. [Google Scholar] [CrossRef]
- Doi, M.; Hirayama, J.; Sassone-Corsi, P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 2006, 125, 497–508. [Google Scholar] [CrossRef]
- Katada, S.; Sassone-Corsi, P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat. Struct. Mol. Biol. 2010, 17, 1414–1421. [Google Scholar] [CrossRef]
- Naruse, Y.; Oh-hashi, K.; Iijima, N.; Naruse, M.; Yoshioka, H.; Tanaka, M. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 2004, 24, 6278–6287. [Google Scholar] [CrossRef]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef]
- Asher, G.; Gatfield, D.; Stratmann, M.; Reinke, H.; Dibner, C.; Kreppel, F.; Mostoslavsky, R.; Alt, F.W.; Schibler, U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008, 134, 317–328. [Google Scholar] [CrossRef]
- Etchegaray, J.P.; Yang, X.; DeBruyne, J.P.; Peters, A.H.; Weaver, D.R.; Jenuwein, T.; Reppert, S.M. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 2006, 281, 21209–21215. [Google Scholar] [CrossRef]
- Ripperger, J.A.; Schibler, U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006, 38, 369–374. [Google Scholar] [CrossRef]
- Raduwan, H.; Isola, A.L.; Belden, W.J. Methylation of histone H3 on lysine 4 by the lysine methyltransferase SET1 protein is needed for normal clock gene expression. J. Biol. Chem. 2013, 288, 8380–8390. [Google Scholar] [CrossRef]
- DiTacchio, L.; Le, H.D.; Vollmers, C.; Hatori, M.; Witcher, M.; Secombe, J.; Panda, S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science 2011, 333, 1881–1885. [Google Scholar] [CrossRef]
- Nam, H.J.; Boo, K.; Kim, D.; Han, D.H.; Choe, H.K.; Kim, C.R.; Sun, W.; Kim, H.; Kim, K.; Lee, H.; et al. Phosphorylation of LSD1 by PKCalpha is crucial for circadian rhythmicity and phase resetting. Mol. Cell 2014, 53, 791–805. [Google Scholar] [CrossRef]
- Jones, M.A.; Covington, M.F.; Ditacchio, L.; Vollmers, C.; Panda, S.; Harmer, S.L. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc. Natl. Acad. Sci. USA 2010, 107, 21623–21628. [Google Scholar] [CrossRef]
- Belden, W.J.; Loros, J.J.; Dunlap, J.C. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell 2007, 25, 587–600. [Google Scholar] [CrossRef]
- Cha, J.; Zhou, M.; Liu, Y. CATP is a critical component of the Neurospora circadian clock by regulating the nucleosome occupancy rhythm at the frequency locus. EMBO Rep. 2013, 14, 923–930. [Google Scholar] [CrossRef]
- Kramer, C.; Loros, J.J.; Dunlap, J.C.; Crosthwaite, S.K. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 2003, 421, 948–952. [Google Scholar] [CrossRef]
- Kunova, A.; Zubko, E.; Meyer, P. A pair of partially overlapping Arabidopsis genes with antagonistic circadian expression. Int. J. Plant Genomics 2012, 2012, 349527. [Google Scholar]
- Smith, K.M.; Sancar, G.; Dekhang, R.; Sullivan, C.M.; Li, S.; Tag, A.G.; Sancar, C.; Bredeweg, E.L.; Priest, H.D.; McCormick, R.F.; et al. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for neurospora white collar complex. Eukaryot. Cell 2010, 9, 1549–1556. [Google Scholar] [CrossRef]
- Dang, Y.; Li, L.; Guo, W.; Xue, Z.; Liu, Y. Convergent transcription induces dynamic DNA methylation at disiRNA loci. PLoS Genet. 2013, 9, e1003761. [Google Scholar] [CrossRef]
- Lewis, Z.A.; Honda, S.; Khlafallah, T.K.; Jeffress, J.K.; Freitag, M.; Mohn, F.; Schubeler, D.; Selker, E.U. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009, 19, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Kouzminova, E.; Selker, E.U. Dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001, 20, 4309–4323. [Google Scholar] [CrossRef]
- Freitag, M.; Hickey, P.C.; Khlafallah, T.K.; Read, N.D.; Selker, E.U. HP1 is essential for DNA methylation in neurospora. Mol. Cell 2004, 13, 427–434. [Google Scholar] [CrossRef]
- Lewis, Z.A.; Adhvaryu, K.K.; Honda, S.; Shiver, A.L.; Knip, M.; Sack, R.; Selker, E.U. DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet. 2010, 6, e1001196. [Google Scholar] [CrossRef]
- Xu, H.; Wang, J.; Hu, Q.; Quan, Y.; Chen, H.; Cao, Y.; Li, C.; Wang, Y.; He, Q. DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet. 2010, 6, e1001132. [Google Scholar] [CrossRef]
- Zhao, Y.; Shen, Y.; Yang, S.; Wang, J.; Hu, Q.; Wang, Y.; He, Q. Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J. Biol. Chem. 2010, 285, 4355–4365. [Google Scholar] [CrossRef]
- Tamaru, H.; Selker, E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 2001, 414, 277–283. [Google Scholar] [CrossRef]
- Tamaru, H.; Zhang, X.; McMillen, D.; Singh, P.B.; Nakayama, J.; Grewal, S.I.; Allis, C.D.; Cheng, X.; Selker, E.U. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003, 34, 75–79. [Google Scholar] [CrossRef]
- Honda, S.; Selker, E.U. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol. Cell. Biol. 2008, 28, 6044–6055. [Google Scholar] [CrossRef]
- Hansen, J. Increased breast cancer risk among women who work predominantly at night. Epidemiology 2001, 12, 74–77. [Google Scholar] [CrossRef]
- Schernhammer, E.S.; Laden, F.; Speizer, F.E.; Willett, W.C.; Hunter, D.J.; Kawachi, I.; Fuchs, C.S.; Colditz, G.A. Night-shift work and risk of colorectal cancer in the nurses' health study. J. Natl. Cancer Inst. 2003, 95, 825–828. [Google Scholar] [CrossRef]
- Davis, S.; Mirick, D.K.; Stevens, R.G. Night shift work, light at night, and risk of breast cancer. J. Natl. Cancer Inst. 2001, 93, 1557–1562. [Google Scholar] [CrossRef]
- Chen, S.T.; Choo, K.B.; Hou, M.F.; Yeh, K.T.; Kuo, S.J.; Chang, J.G. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis 2005, 26, 1241–1246. [Google Scholar] [CrossRef]
- Taniguchi, H.; Fernandez, A.F.; Setien, F.; Ropero, S.; Ballestar, E.; Villanueva, A.; Yamamoto, H.; Imai, K.; Shinomura, Y.; Esteller, M. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 2009, 69, 8447–8454. [Google Scholar] [CrossRef]
- Hsu, M.C.; Huang, C.C.; Choo, K.B.; Huang, C.J. Uncoupling of promoter methylation and expression of Period1 in cervical cancer cells. Biochem. Biophys. Res. Commun. 2007, 360, 257–262. [Google Scholar] [CrossRef]
- Yang, M.Y.; Chang, J.G.; Lin, P.M.; Tang, K.P.; Chen, Y.H.; Lin, H.Y.; Liu, T.C.; Hsiao, H.H.; Liu, Y.C.; Lin, S.F. Downregulation of circadian clock genes in chronic myeloid leukemia: Alternative methylation pattern of hPER3. Cancer Sci. 2006, 97, 1298–1307. [Google Scholar] [CrossRef]
- Shih, M.C.; Yeh, K.T.; Tang, K.P.; Chen, J.C.; Chang, J.G. Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol. Carcinog. 2006, 45, 732–740. [Google Scholar] [CrossRef]
- Yeh, K.T.; Yang, M.Y.; Liu, T.C.; Chen, J.C.; Chan, W.L.; Lin, S.F.; Chang, J.G. Abnormal expression of period 1 (PER1) in endometrial carcinoma. J. Pathol. 2005, 206, 111–120. [Google Scholar] [CrossRef]
- Zhu, Y.; Stevens, R.G.; Hoffman, A.E.; Tjonneland, A.; Vogel, U.B.; Zheng, T.; Hansen, J. Epigenetic impact of long-term shiftwork: Pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol. Int. 2011, 28, 852–861. [Google Scholar] [CrossRef]
- Lee, H.C.; Li, L.; Gu, W.; Xue, Z.; Crosthwaite, S.K.; Pertsemlidis, A.; Lewis, Z.A.; Freitag, M.; Selker, E.U.; Mello, C.C.; et al. Diverse pathways generate microRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol. Cell 2010, 38, 803–814. [Google Scholar] [CrossRef]
- Li, N.; Joska, T.M.; Ruesch, C.E.; Coster, S.J.; Belden, W.J. The frequency natural antisense transcript first promotes, then represses, frequency gene expression via facultative heterochromatin. Proc. Natl. Acad. Sci. USA 2014. submitted. [Google Scholar]
- Ruesch, C.E.; Chong, H.S.; Park, J.; Li, N.; Joska, T.M.; Belden, W.J. The histone H3 lysine 9 methylastransferase DIM-5 modifies chromatin at frequency and represses light-activated gene expression. G3 2014. submitted. [Google Scholar]
- Lippman, Z.; May, B.; Yordan, C.; Singer, T.; Martienssen, R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003, 1, E67. [Google Scholar] [CrossRef] [Green Version]
- Soppe, W.J.; Jasencakova, Z.; Houben, A.; Kakutani, T.; Meister, A.; Huang, M.S.; Jacobsen, S.E.; Schubert, I.; Fransz, P.F. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002, 21, 6549–6559. [Google Scholar] [CrossRef]
- Tariq, M.; Saze, H.; Probst, A.V.; Lichota, J.; Habu, Y.; Paszkowski, J. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 2003, 100, 8823–8827. [Google Scholar] [CrossRef]
- Fu, L.; Pelicano, H.; Liu, J.; Huang, P.; Lee, C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002, 111, 41–50. [Google Scholar] [CrossRef]
- Miki, T.; Matsumoto, T.; Zhao, Z.; Lee, C.C. P53 regulates Period2 expression and the circadian clock. Nat. Commun. 2013, 4, 2444. [Google Scholar] [CrossRef]
- Hill, S.M.; Blask, D.E.; Xiang, S.; Yuan, L.; Mao, L.; Dauchy, R.T.; Dauchy, E.M.; Frasch, T.; Duplesis, T. Melatonin and associated signaling pathways that control normal breast epithelium and breast cancer. J. Mammary Gland Biol. Neoplasia 2011, 16, 235–245. [Google Scholar] [CrossRef]
- Sarkar, D.K.; Zhang, C.; Murugan, S.; Dokur, M.; Boyadjieva, N.I.; Ortiguela, M.; Reuhl, K.R.; Mojtehedzadeh, S. Transplantation of beta-endorphin neurons into the hypothalamus promotes immune function and restricts the growth and metastasis of mammary carcinoma. Cancer Res. 2011, 71, 6282–6291. [Google Scholar] [CrossRef]
- Xie, C.; Yuan, J.; Li, H.; Li, M.; Zhao, G.; Bu, D.; Zhu, W.; Wu, W.; Chen, R.; Zhao, Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014, 42, D98–D103. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Joska, T.M.; Zaman, R.; Belden, W.J. Regulated DNA Methylation and the Circadian Clock: Implications in Cancer. Biology 2014, 3, 560-577. https://0-doi-org.brum.beds.ac.uk/10.3390/biology3030560
Joska TM, Zaman R, Belden WJ. Regulated DNA Methylation and the Circadian Clock: Implications in Cancer. Biology. 2014; 3(3):560-577. https://0-doi-org.brum.beds.ac.uk/10.3390/biology3030560
Chicago/Turabian StyleJoska, Tammy M., Riasat Zaman, and William J. Belden. 2014. "Regulated DNA Methylation and the Circadian Clock: Implications in Cancer" Biology 3, no. 3: 560-577. https://0-doi-org.brum.beds.ac.uk/10.3390/biology3030560



