1. Introduction
Amaranthus L. is a genus in the family Amaranthaceae consisting of more than 60 species, including cultivated grain amaranth species (e.g.,
A. caudatus, A. cruentus, and
A. hypochondriacus) and leaf vegetable types (e.g.,
A. hybridus and
A. tricolor) [
1,
2]. They are relatively short-lived annuals that grow vigorously, are drought resistant and adapt readily to new environments [
3]. The major centers of distribution are tropical America, India, China, Nepal, Italy, Greece, Africa, and Australia [
4,
5]. Amaranths are ancient crops with promising economic and nutritional value and have attracted increasing interest over recent decades [
6,
7,
8]. Grain amaranth has been gaining worldwide acceptance as a crop rich in high-quality protein (13%–19% of dry weight) and its remarkable essential amino acid balance which is close to the optimum protein reference pattern in the human diet according to FAO/WHO requirements [
9,
10,
11]. Grain amaranth is also an excellent source of minerals, vitamins A, C, and E [
12,
13,
14]. Considering its agronomic and nutritional importance, amaranth has potential to broaden the food base in sub-Saharan Africa [
15]; therefore, attention should be given to the cultivation, genetic improvement, and sustainable utilization of this promising crop in the region.
Amaranth is a self-pollinated crop, but wide variation in genotypes exists due to varying amounts of outcrossing and frequent interspecific and inter-varietal hybridization [
1,
16]. Amaranths also exhibit tremendous diversity related to their wide adaptability to different eco-geographic situations [
17]. Genotype identification in amaranth had been a long term challenge [
18] due to the close relationships that exists within the genus.
Knowledge of genetic diversity and trait variations in populations is useful in plant breeding and for developing
ex situ conservation strategies of plant genetic resources [
19,
20,
21]. Plant phenotype is commonly used for estimating genetic diversity as it provides a simple way of quantifying genotypic variation. However, the efficiency of estimating genetic differences using morphological traits is largely limited by uncontrollable environmental factors and distinctness. Molecular-based analyses overcome many limitations of morphological and biochemical trait-based procedures and have been used variously in determining genetic diversity [
22,
23]. Various types of molecular markers are utilized to evaluate DNA polymorphism. Random Amplified Polymorphic DNA (RAPD), a PCR (polymerase chain reaction)-based technique has considerable advantage for studying plant genome characterizations because it is simple, relatively inexpensive, utilizes arbitrary primers, and randomly samples a potentially large number of loci in a less complex pattern [
1,
24,
25]. Even though RAPD is criticized for low reproducibility, this is overcome by optimization of the reaction and maintenance of stringent conditions [
26,
27]. RAPD analyses has been employed for several plant species in relation to development of genetic conservation and improvement strategies, including the study of genetic diversity, taxonomic delimitations, and evolutionary relationships in
Amaranthus species [
1,
17,
28,
29].
Grain amaranth remains a largely underexploited crop for grain purposes in sub-Saharan Africa despite the potential as a hardy field crop with generally excellent nutritional qualities. However, research aimed at its genetic improvement and agronomic adoption in sub-Saharan Africa is almost non-existent. A number of accessions of amaranth have been introduced that have acclimatized well in the region, but evaluation of agronomic and nutritional traits in the peculiar agro-ecologies of the region has not been conducted. The availability of genetic variation among and within the different accessions for these traits provides great scope for crop improvement through selection and other breeding methods to develop desired genotypes. Thus, insight into the genetic variation within and among the available amaranth genotypes in relation to the morphological and nutritional traits as revealed by RAPD markers is necessary. Such empirical knowledge will facilitate strategic marker-assisted selection (MAS) breeding, as well as enhance effective genetic resources exploration, conservation, management, and utilization of Amaranthus species in future breeding programs.
The aim of this work was therefore to study the genetic variation observable in 29 Amaranthus accessions in the agro-ecology of South West Nigeria using phenotypic traits of agronomic and nutritional significance, and RAPD markers. The goal is to provide an insight for further utilization of RAPD markers to characterize and identify quantitative trait loci (QTLs) for agronomic and nutritional quality in amaranth genotypes bred for adoption in sub-Saharan agro-ecologies. Subsequently, RAPDs adjacent to important QTLs will be used in marker-assisted selection (MAS) of breeding lines.
4. Discussion and Conclusions
Grain cultivars of
Amaranthus species are cultivated in many regions of the world for the seeds which are rich in protein, vitamins, and mineral components [
9,
11,
14]. In recent years, a lot of emphasis has been made on expanding their cultivation; however, seed yields may be low when grown outside environments for which the particular genotype was bred. This could largely limit the crop’s use for subsistence, as well as commercial cultivation in less-than-optimum environments such as sub-Saharan Africa. In view of the enormous nutritional benefits of the crop, there is a definite need for its genetic improvement to develop high yielding varieties with high content of desired nutritional traits that are well adapted to the agro-ecological climates of sub-Saharan Africa. In the process of genetic improvement of any crop, genetic diversity among germplasm plays a major role, since it opens the way to determine the most divergent parents based on contribution of different qualitative and quantitative traits for utilization in breeding programs.
The traits investigated in this study are important, as they have direct and indirect effects on seed yield and its nutritional components. Studies on vegetable amaranth showed the presence of wide range of diversity in both agronomic and qualitative traits especially leaf and stem color [
38,
39]. Xiao
et al. [
39] further opined that stem and leaf colour were useful indices in the classification of vegetable amaranths. Wu
et al. [
38] in a study of 229 genotypes from 20
Amaranthus species observed wide variability which was useful in cultivar improvement for agronomic traits such as plant height, seed yield, stem, and leaf color among genotypes. Similarly in this study, characters such as plant height, stem diameter, number of leaves per plant, leaf area, and seed yield had high coefficients of variation indicating scope for improvement in these traits through selection to enhance the potentiality of seed yield and plant vigor. Results from traits, such as days to 50% flowering, moisture content, starch, fat, protein, magnesium, manganese, and phosphorus had low values of coefficient of variation, which implies that chances of getting substantial gains under selection are likely to be less for these traits. Additionally, the negative correlation between plant height and number of leaves and branches is an indication that most of the tall accessions have fewer numbers of leaves and branches.
In this study, the PCA showed that the first four PCs accounted for about 57.53% of the total variation encountered among the accessions taking into account all the 27 traits simultaneously. PC
1 distinguished those accessions that gave high plant height, number of branches, stem diameter, leaf area, fat, zinc, and copper content. PC
2 distinguished accessions having high values of number of leaves and most of the mineral contents (iron, aluminum, magnesium, potassium, selenium, phosphorus, and calcium). Although calcium content had some strength in PC
2, it appeared more strongly in PC
4. Selenium content was prominent in PC
3, but appeared more strongly in PC
2. This may be an indication that calcium and selenium are not strongly correlated with the rest of the traits and, hence, it could be possible to select for genotypes with high calcium and selenium content without necessarily affecting other economically important traits. PC
3 distinguished accessions having high values of sugar, starch, crude fiber, and selenium content, while PC
4 distinguished accessions having high seed yield, 100-seed weight, moisture, ash, protein, manganese, and calcium content. Protein has been reported to be a major contributing trait of PC
2 in
Amaranthus [
2,
40]; in this study protein is a major contributing trait of PC
4.
The germplasm under evaluation was grouped into five clusters on the basis of phehotypic traits. Accessions originating from the same region more or less clustered together in some cases, as was observed in Cluster III with accessions A16, A17, A18, and A19, which are from Oyo State in Nigeria. This may suggest that certain traits could be more confined to certain geographical regions than in others. Shukla
et al. [
2] reported a similar trend in strains of vegetable amaranth of the Tarai region and of the Himalayan region being segregated into same clusters. The clustering together of amaranth accessions from Oyo State of Nigeria indicates that they may be adapted to the agro-ecological conditions characteristic of the region. Accessions A5, A6, A7, A8, A9, A10, and A12 belonging to
A. cruentus were also grouped together in cluster III. Thus, the
Amaranthus hybrid aceessions and those of
A. cruentus may be closely related in terms of phenotypic characteristics as they turned up together in cluster III having high protein and selenium contents. Accessions showing high mean performance for specific desired traits can be used as donor parents in hybridization for improving those traits in component breeding [
41]. Cluster IV with only one accession, A11 (
A. cruentus), was tall, thick stemmed, had high leaf area, seed yield, fat, and some microelements. Similarly, Cluster V also had only one accession, A23 (
A. hypochondriacus), which had high leaf number, seed weight and most of the macroelements. These accessions can be considered as sources of genes for seed yield, plant vigor, and mineral elements in amaranth improvement programs. Ugborogho and Oyelana [
42] observed in their investigations that many accessions of
A. hybridus exist and they tend to vary in plant height, leaf, stem and seed inflorescence color, and grain yield. In this study,
A. hybridus (A28 and A29) in Clusters II and III can be good sources of genes for starch, protein, and selenium content since they are high in these traits.
The result of cluster analysis is consistent with the results obtained through PCA, whereby the major differences between the clusters were attributed to the same traits that contributed most to PC
1, PC
2, and PC
3. As similar result was observed in genotypic diversity studies on Ethiopian mustard [
43],
Amaranthus tricolor [
2] and
Amaranthus hybridus [
44]. Findings from this study are in concordance with earlier reports that both PCA and cluster analysis can disclose complex relationships between taxa in a more understandable way and with equal effectiveness [
45,
46].
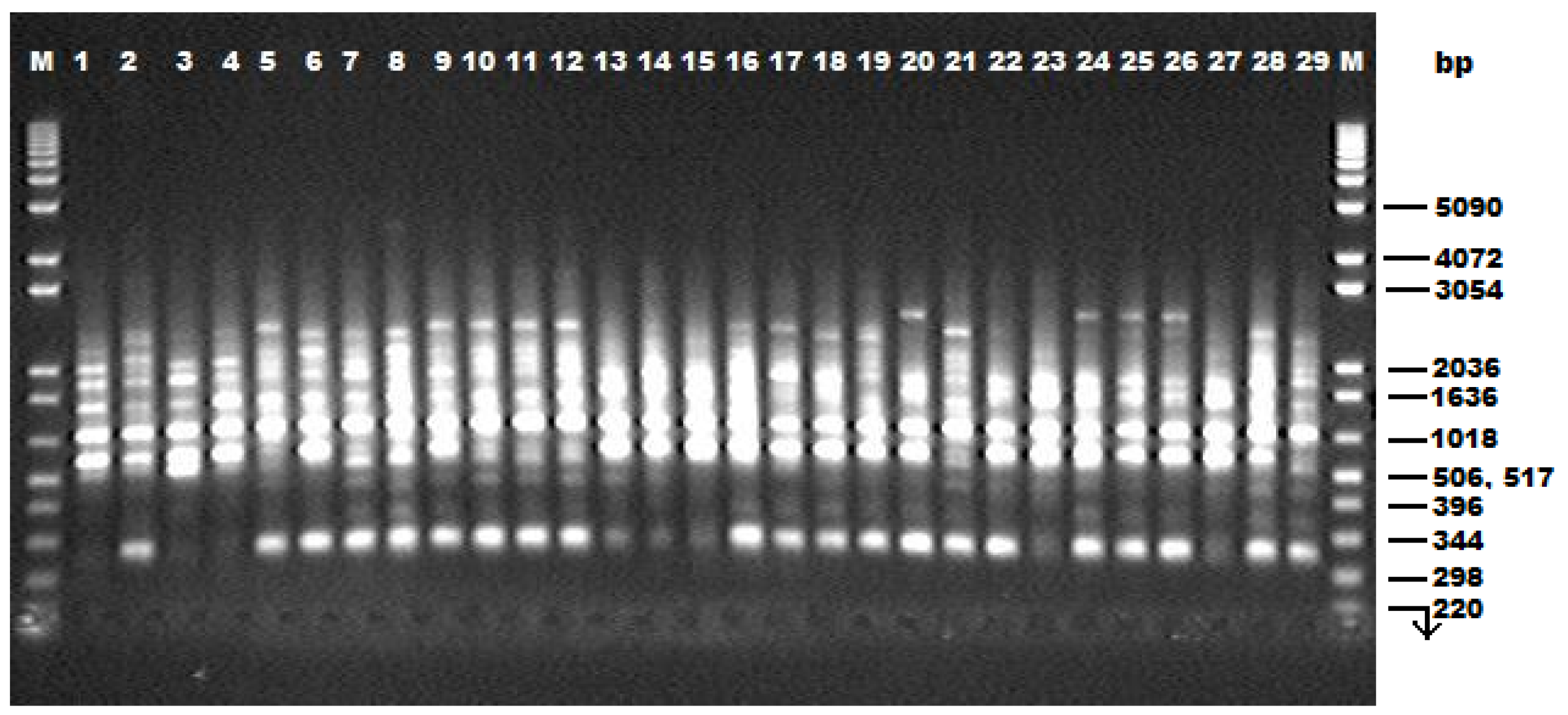
The amaranth germplasm collection was also assessed for diversity at DNA level. Only 16 of 40 RAPD primers screened gave amplification. These generated 193 fragments of which 157 were polymorphic (81%). Pejie
et al. [
47] reported that 150 polymorphic bands should make a reliable estimate of genetic similarities possible among genotypes within the same species. Hence, the number of polymorphic information produced by the 16 RAPD primers in this study can give an estimate of the genetic diversity in grain amaranth. The range of genetic diversity values broadly indicates the degree of heterogeneity or homogeneity in different accessions of the plant species [
48]. Study on genetic diversity of 16
Butea monosperma accessions representing four agro-ecological zones from nine sub-climatic regions in India using 12 polymorphic RAPD primers generated a similarity matrix based on Jaccard’s coefficient ranging from 0.53 to 0.79. This low range of genetic diversity may imply conservation of germplasm and low level of heterogeneity [
49]. In this study the 29 amaranth accessions which were of different geographical origins had genetic similarity coefficients of between 0.61 and 0.89. At the intraspecific level, percentage of RAPD polymorphism has previously been reported to be 22.5%, 18.3%, and 23.3% for
A. hypochondriacus,
A. caudatus, and
A. cruentus [
1]. Chan and Sun [
29] reported a higher percentage of polymorphism in leafy amaranths at the intraspecific level than in grain amaranths. In this study, the high percentage of intraspecific polymorphism observed in
A. hypochondriacus (66.32%) and in
A. hybrid (61.14%) indicates that they both sustain greater genetic variation than the other amaranth species studied (
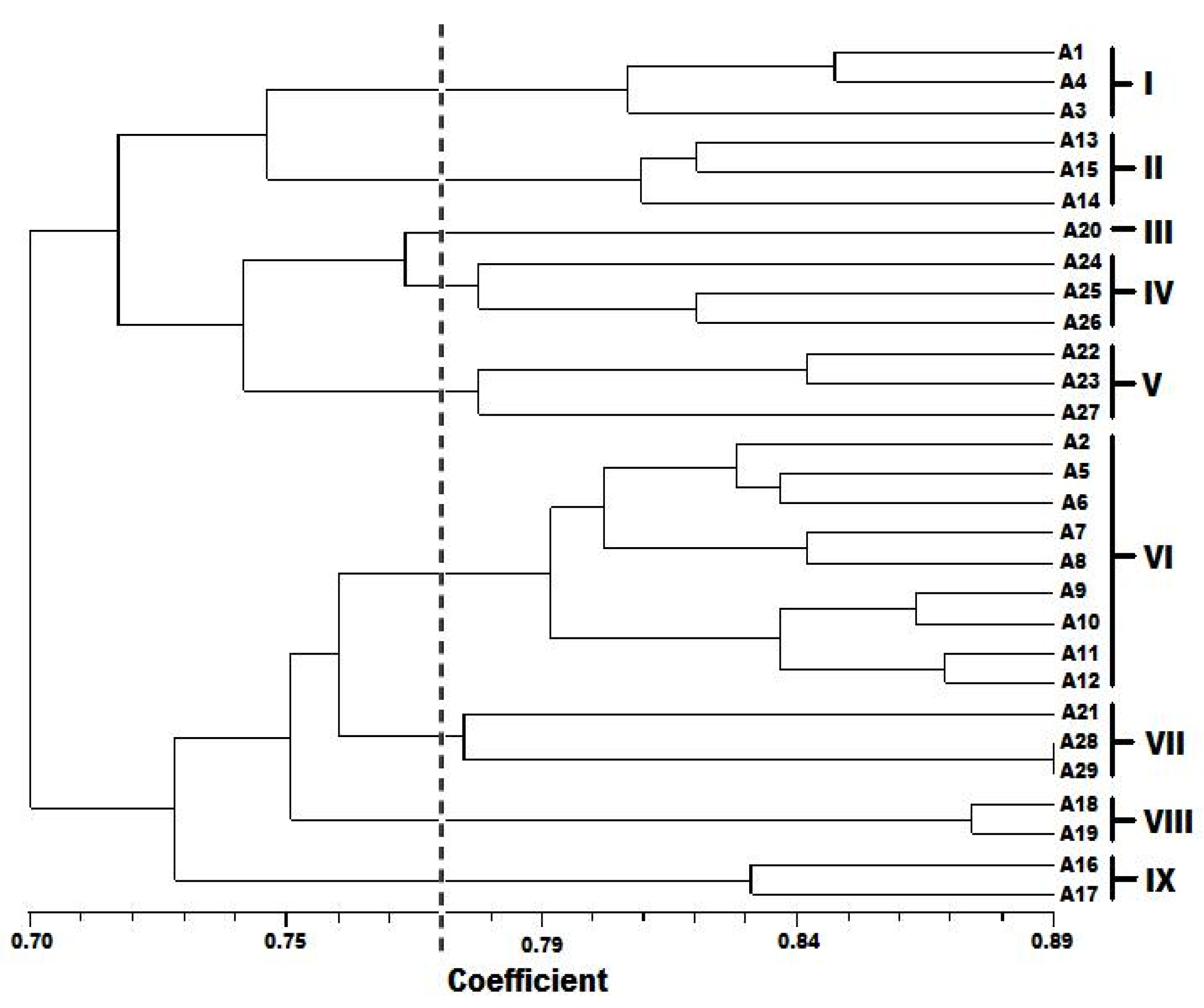
Table 7). Most populations of a species clustered together as observed in the dendrogram generated by UPGMA (
Figure 3), except for accessions A2, A20, A21, and A27 which were found clustering with other species. It is plausible that the primers used in this study amplified mostly the conserved part of the genome, so they could not show any variation within a population. Of the 29 accessions analyzed, two accessions, A28 (NH84/444-4) and A29 (NHAC3) belonging to
A. hybridus from Oyo, Nigeria; displayed the maximum genetic similarity with a 0.89 similarity coefficient value. It is also evident from the RAPD-based clustering that
A. caudatus is closely related to
A. hybrid and
A. hypochondriacus. Similar results were also observed in previous RAPD based analyses on
Amaranthus [
1,
29,
50]. This study revealed low level of diversity as can be observed in the dendrogram showing about 70% similarity and 30% diversity in the accessions.
This study showed some extent of geographic cohesiveness, as observed in accessions A13, A14, and A15; A16 and A17; and A18 and A19. These clustered together in clusters II, IX, and VIII, respectively (
Figure 3). Similar observations were made by Ray and Roy [
1] when they noted similar geographic and ecological patterns in the distribution of genetic diversity among populations of
Amaranthus species from the Indo-Gangetic plains. Other studies have showed little relationship [
51], whereas some clearly demonstrated noticeable associations between population characteristics and the environment in which they occur [
52].
The study of genetic diversity among populations of different phytogeographic regions is important, as survival, perpetuation, and continuance of a species to meet the demands of changing environments largely depends on the extent of variability available in its gene pool. Selection in genotypes amaranth species should be based mainly on some distinctive morphological characters, such as plant height, stem diameter, number of leaves, number of branches, leaf area, and seed yield. Accessions A11 (PI 641047), A23 (PI 590991) and A28 (NH 84/444-4), having high seed yield, protein, crude fiber, and essential minerals could serve as promising sources of genes for these nutrients in future hybridization work. The RAPD analysis clearly revealed genetic diversity among and within accessions of the species studied, showing that this DNA marker is a useful tool not only for assessing intraspecific variation, but also for subsequent characterization and identification of quantitative trait loci (QTLs) for agronomic and nutritional quality in amaranth genotypes specifically bred for sub-Saharan agro-ecologies.