Exerting DNA Damaging Effects of the Ilimaquinones through the Active Hydroquinone Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
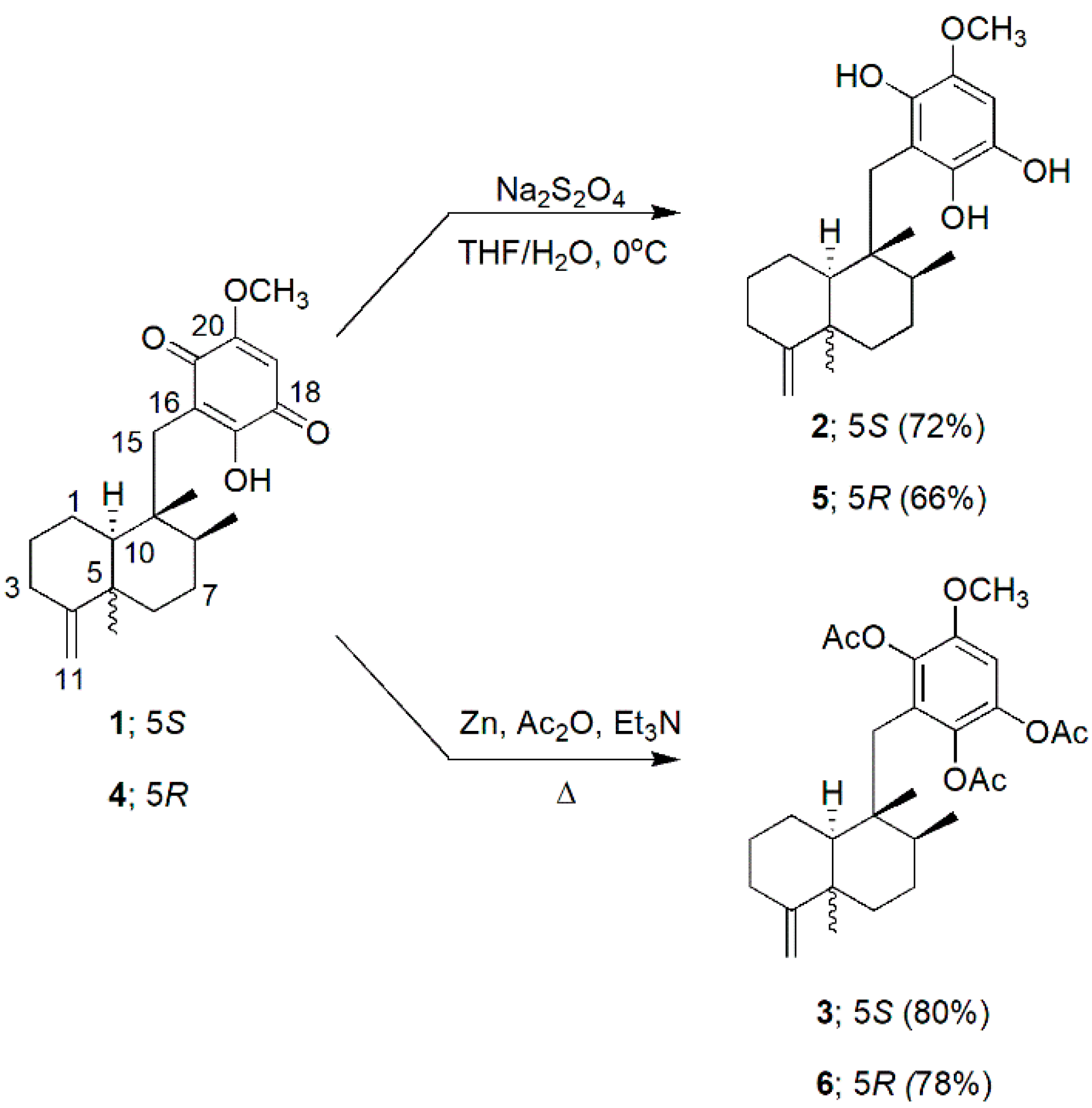
2.2. Dithioite Reduction and Reductive Peracetylation of 1 and 4
2.2.1. Dithionite Reduction of 1
2.2.2. Reductive Peracetylation of 1
2.2.3. Dithionite Reduction of 4
2.2.4. Reductive Peracetylation of 4
2.3. Cytotoxic Activity Determinaiton
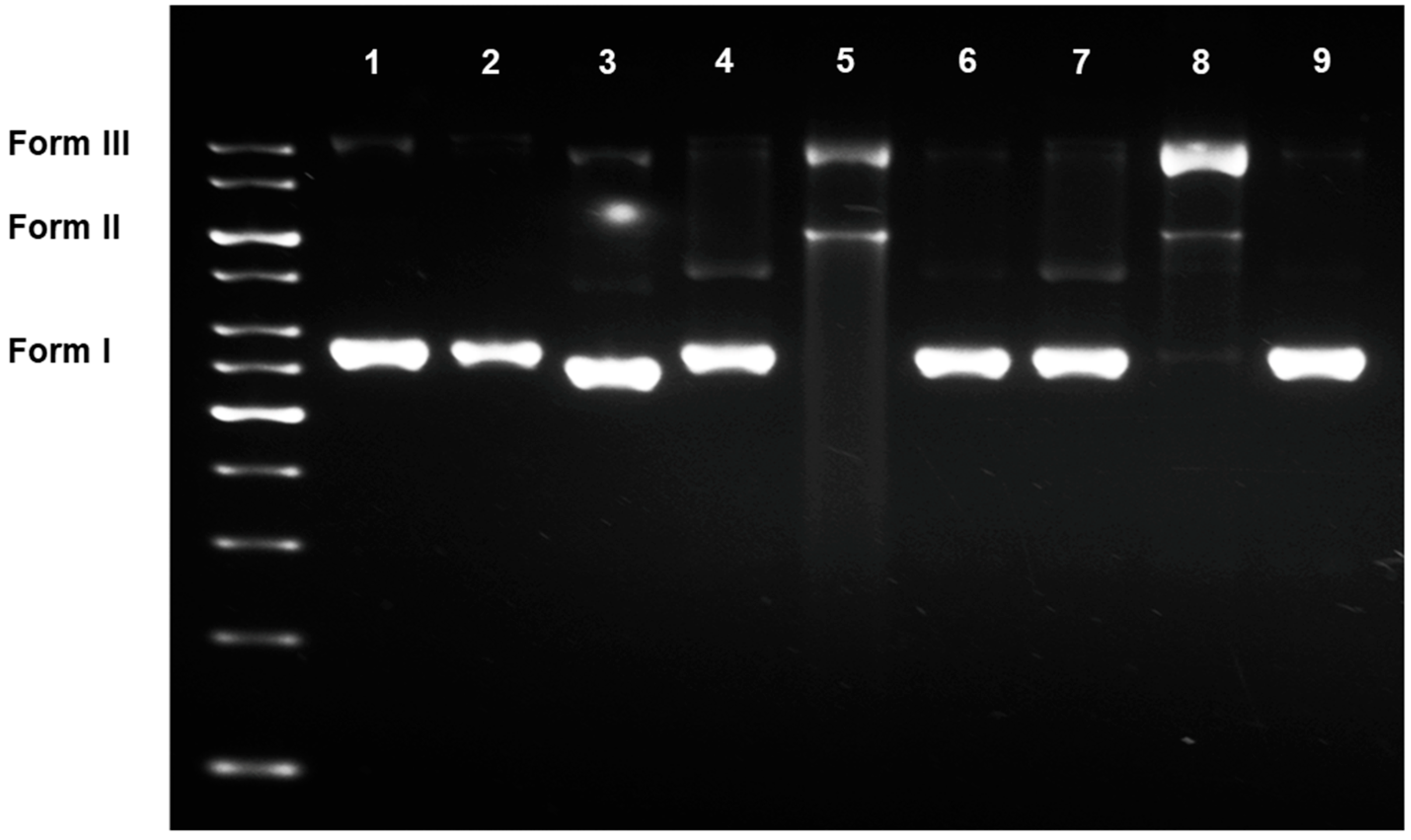
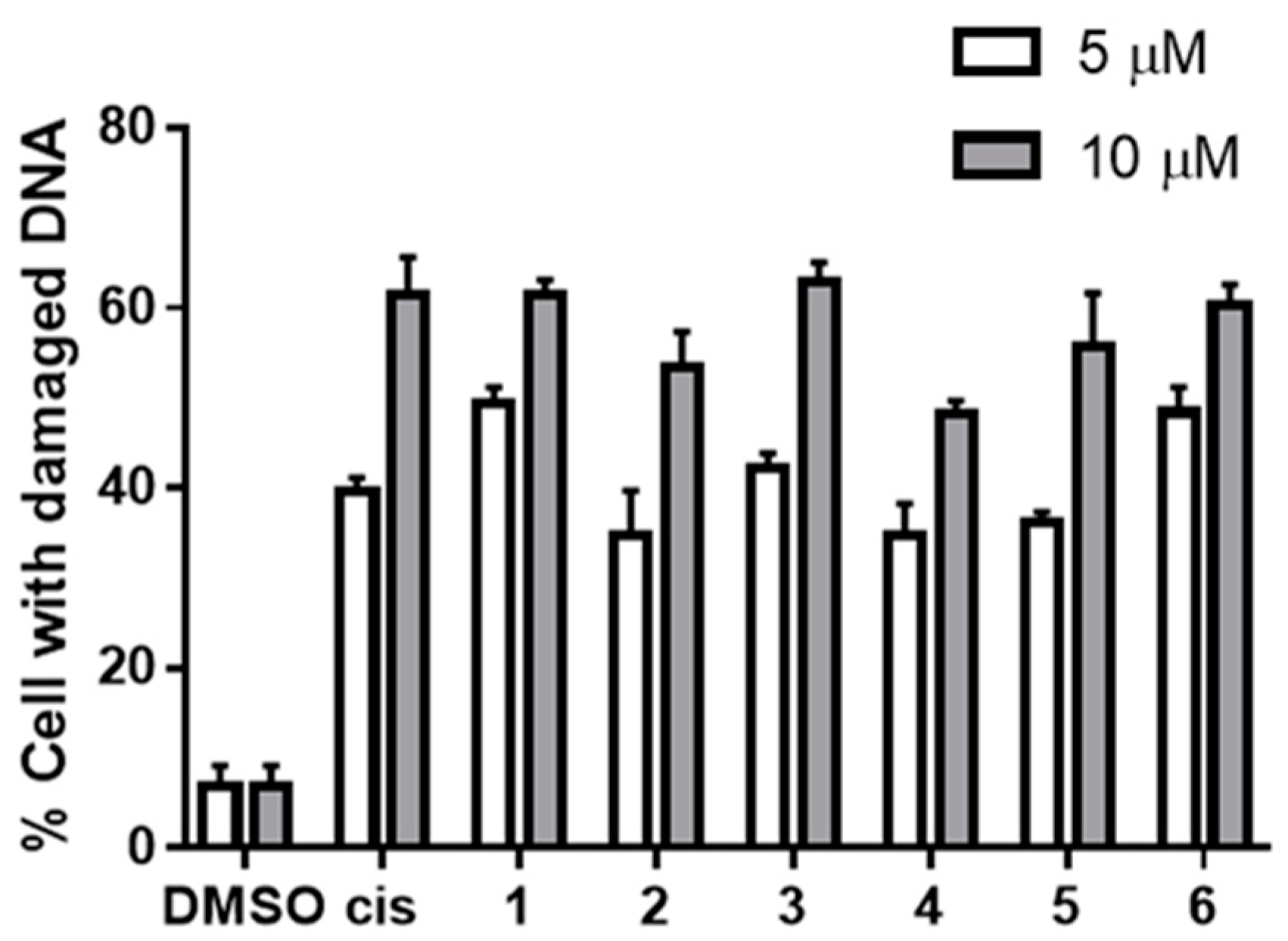
2.4. DNA Damaging Experiments and Agarose Gel Electrophoreses
2.5. Hoechst 33342 Nuclear Staining
2.6. NQO1 Inhbiting Assays
3. Results and Discussion
3.1. Preparation of Hydroquinones and Hydroquinone Triacetates
3.2. Cytotoxic Activities and DNA Damaging Effects of Ilimaquinone and Derivatives
3.2.1. Cytotoxic Activities
3.2.2. DNA Damaging Effects in a Cell-Free System
3.2.3. DNA Damaging Effects in a Cell-Based Assay
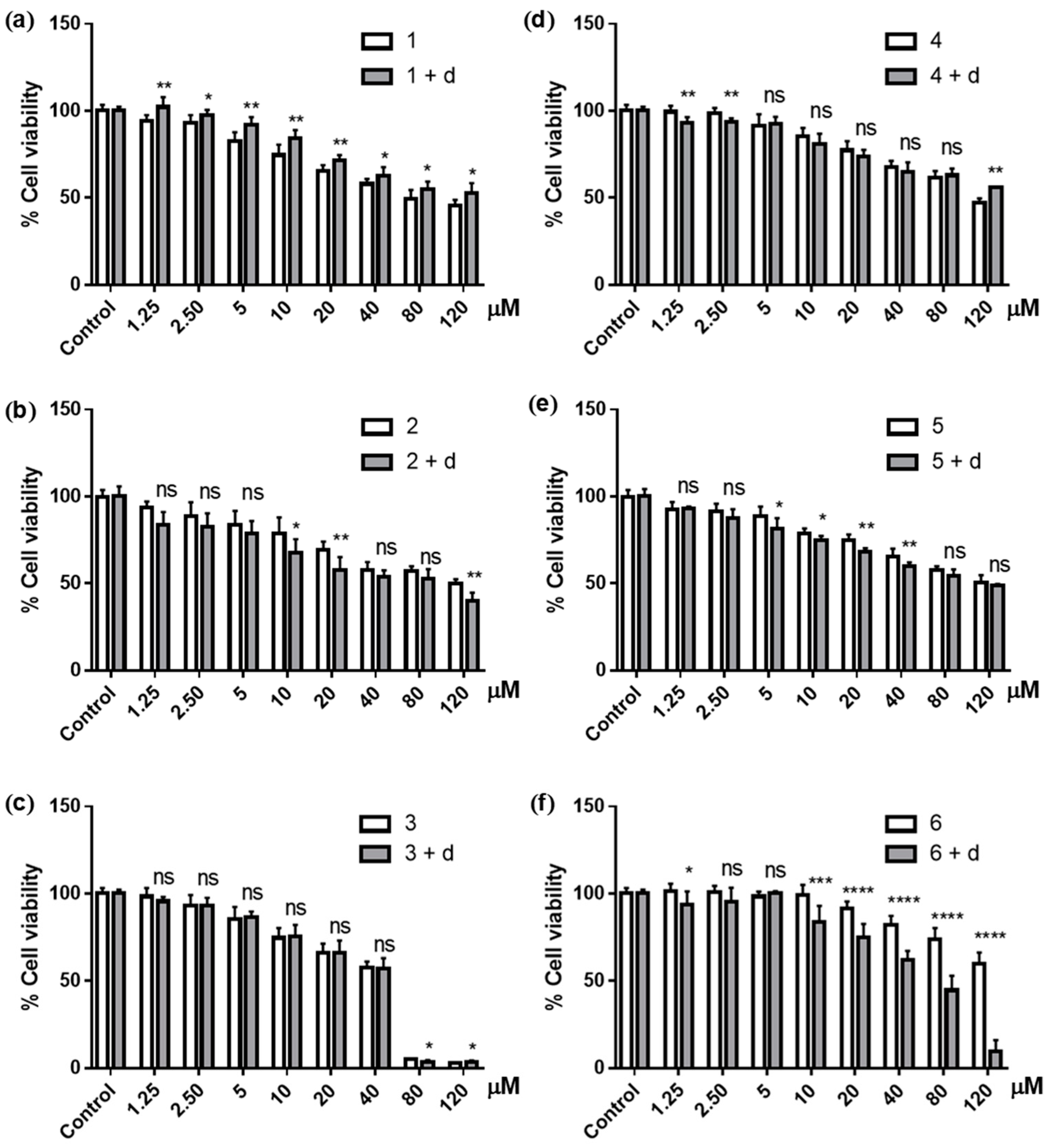
3.3. Effects of NQO1 Inhibition on the Cytotoxicity of Ilimaquinone Derivatives
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, G.S.; Lipman, R.; Cummings, J.; Tomasz, M. Mitomycin C-DNA adducts generated by DT-diaphorase. Revised mechanism of the enzymatic reduction activation of mitomycin C. Biochemistry 1997, 36, 14128–141360. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, P.L. Mechanism(s) of bioreductive activation: The example of diaziquinone (AZQ). Free Radic. Biol. Med. 1989, 6, 405–445. [Google Scholar] [CrossRef]
- Balansa, W.Q.; Mettal, U.; Wusam, Z.G.; Plubrukarn, A.; Ijong, F.G.; Liu, Y.; Schäberle, T.F. A new sesquiterpene aminoquinone from an Indonesian marine sponge. Mar. Drugs 2019, 17, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiso, A.; Kittiwisut, S.; Chantakul, R.; Yuenyongsawad, S.; Putchakarn, S.; Schäberle, T.F.; Temkitthaworn, P.; Ingkaninan, K.; Chaithirayanon, K.; Plubrukarn, A. Quintaquinone, a merosesquiterpene from the yellow sponge Verongula cf. rigida Esper. J. Nat. Prod. 2020, 82, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.H.; Chueh, S.C.; Kung, F.L.; Pan, S.L.; Shen, Y.C.; Guh, J.H. Ilimaquinone, a marine sponge metabolite, display anticancer activity via GADD153-mediated pathway. Eur. J. Pharmacol. 2007, 556, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, G.; Namikoshi, M.; Kobayashi, H.; Yao, X.; Cai, G. Sesquiterpene quinones from a marine sponge Hippospongia sp. that inhibit maturation of starfish oocytes and induce cell cycle arrest with HepC2 cells. Pharm. Biol. 2006, 76, 522–527. [Google Scholar] [CrossRef]
- Du, L.; Zhoa, Y.D.; Nagle, D.G. Inducers of hypoxic response: Marine sesterterpene quinones activiate HIF-1. J. Nat. Prod. 2013, 76, 1175–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Yun, E.; Hwang, H.I.; Yoon, S.; Kim, D.E.; Kim, S.J.; Na, M.; Song, G.Y.; Oh, S. Ilimaquinone and ethylmenoquionone, marine sponge metabolites, suppress the proliferation of multiple myeloma cells by down-regulating the level of β catenin. Mar. Drugs 2014, 12, 3231–3244. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Chung, J.K.; Hwang, H.I.; Gwak, J.; Park, S.; Ju, G.B.; Yun, E.; Kim, D.E.; Chung, Y.H.; Na, M.; et al. Activation of p53 with ilimaquinone and ethylmenoquinone, marine sponge metabolites, induce apoptosis and autophagy in colon cancer cells. Mar. Drugs 2015, 13, 543–557. [Google Scholar] [CrossRef] [Green Version]
- Ratovitski, A.E. Tumor protein (TP)-p53 members as regulators of autophagy in tumor cells upon marine drug exposure. Mar. Drugs 2016, 14, 154. [Google Scholar] [CrossRef] [Green Version]
- Müller, W.E.G.; Sladić, D.; Zahn, R.K.; Bässler, K.J.; Dogović, N.; Gerner, H.; Gašić, M.J.; Schröder, H.C. Avarol-induced DNA strand breakage in vitro and in erythroleukemia cells. Cancer Res. 1987, 24, 6565–6571. [Google Scholar]
- Watson, A.T.; Park, K.; Wiemer, D.F.; Scott, W.J. Application of the nickel mediated neopentyl coupling in the total synthesis of the marine natural products arenarol. J. Org. Chem. 1995, 60, 5102–5106. [Google Scholar] [CrossRef]
- Luibrand, R.T.; Erdman, T.R.; Vollmer, J.J.; Scheuer, P.J.; Finer, J.; Clardy, J. Ilimaquinone, a sesquiterpene quinone from a marine sponge. Tetrahedron 1979, 35, 609–612. [Google Scholar] [CrossRef]
- Mosman, T. Rapid colorimetric assay for cellylar growth and survival: Application to proliferation and cytotoxic assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Crowley, L.C.; Marfell, B.J.; Waterhouse, N.J. Analyzing cell death by nuclear staining with Hoechst 33342. Cold Spring Harb. Protoc. 2016, 9, 778–781. [Google Scholar] [CrossRef] [PubMed]
- Ernster, L. DT diaphorase. Methods Enzymol. 1967, 10, 309–317. [Google Scholar]
- Ross, D.; Siegel, D.; Beall, H.; Prakash, A.S.; Mulcahy, R.T.; Gibson, N.W. DT-diaphorase in activation and detoxification of quinones. Cancer Metastasis Rev. 1993, 12, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.T.; Park, H.J. Implication of NQO1 in cancer therapy. BMB Rep. 2015, 48, 609–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Cullen, J.J.; Hinkhouse, M.M.; Grady, M.; Gaut, A.W.; Liu, J.; Zhang, Y.P.; Weydert, C.J.D.; Domann, F.E.; Oberley, L.W. Dicumarol inhibition of NADPH:quinone oxidoreductase induces growth inhibition of pancreatic cancer via a superoxide-mediated mechanisms. Cancer Res. 2003, 63, 5513–5520. [Google Scholar] [PubMed]
- Brunmark, A.; Cadenas, E. Redox and addition chemistry of quinoid compounds and its biological implications. Free Radic. Biol. Med. 1989, 7, 435–477. [Google Scholar] [CrossRef]
- Popov, A.M.; Stekhova, S.I.; Utkina, N.K.; Rebachuk, A.M. Antimicrobial and cytotoxic activity of sesquiterpene quinones and brominated diphenyl esters isolated from marine sponges. Pharm. Chem. J. 1999, 33, 71–73. [Google Scholar] [CrossRef]
- Evanno, L.; Lachkar, D.; Lamali, A.; Boufridi, A.; Séon-Méniel, B.; Tintillier, F.; Saulnier, S.; Denis, S.; Genta-Jouve, G.; Jullian, J.-C.; et al. A ring distortion strategy from marine natural product ilimaquinone leads to quorum sensing modulators. Eur. J. Org. Chem. 2018, 2018, 2486–2497. [Google Scholar] [CrossRef]
- Collier, A.C.; Pritsos, C.A. The mitochondrial uncoupler dicumarol disrupts the MTT assay. Biochem. Pharmacol. 2003, 66, 281–287. [Google Scholar] [CrossRef]
- Siegel, D.; Ross, D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic. Biol. Med. 2000, 29, 246–253. [Google Scholar] [CrossRef]
- Watanabe, J.; Nishiyama, H.; Matsui, Y.; Ito, M.; Kawanishi, H.; Kamoto, T.; Ogawa, O. Dicoumarol potentiates cisplatin-induced apoptosis mediated by c-Jun N-terminal kinase in p53 wild-type urogenital cancer cell lines. Oncogene 2006, 25, 2500–2508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Stuijvenberg, J.; Proksch, P.; Fritz, G. Targeting the DNA damage response (DDR) by natural compounds. Bioorganic Med. Chem. 2020, 28, 115279. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Bai, L.Y.; Su, J.H.; Chiu, C.F.; Lin, W.Y.; Huang, W.T.; Shih, M.C.; Huang, Y.T.; Hu, J.L.; Weng, J.R. Ilimaquinone induces apoptosis and autophagy in human oral squamous cell carcinoma cells. Biomedicines 2020, 8, 296. [Google Scholar] [CrossRef] [PubMed]


| Compounds | IC50 ± SD (μM) |
|---|---|
| 1 | 10.1 ± 2.02 |
| 2 | 17.4 ± 1.11 |
| 3 | 15.7 ± 1.08 |
| 4 | 21.5 ± 1.12 |
| 5 | 17.5 ± 1.11 |
| 6 | 16.6 ± 1.07 |
| docetaxel | 2.0 ± 1.01 |
| cisplatin | 31.2 ± 1.20 |
| Compounds | IC50 ± SD (μM) | |
|---|---|---|
| Without Dicoumarol | Pretreated with Dicoumarol | |
| 1 | 72.6 ± 1.07 | 109.2 ± 1.09 |
| 2 | 86.3 ± 1.13 | 62.6 ± 1.13 |
| 3 | 51.8 ± 1.10 | 51.7 ± 1.10 |
| 4 | 116.5 ± 1.07 | 153.4 ± 1.14 |
| 5 | 126.2 ± 1.09 | 100.8 ± 1.07 |
| 6 | 99.0 ± 1.17 | 51.3 ± 1.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiso, A.; Yurasakpong, L.; Janta, S.; Chaithirayanon, K.; Plubrukarn, A. Exerting DNA Damaging Effects of the Ilimaquinones through the Active Hydroquinone Species. Sci. Pharm. 2021, 89, 26. https://0-doi-org.brum.beds.ac.uk/10.3390/scipharm89020026
Jiso A, Yurasakpong L, Janta S, Chaithirayanon K, Plubrukarn A. Exerting DNA Damaging Effects of the Ilimaquinones through the Active Hydroquinone Species. Scientia Pharmaceutica. 2021; 89(2):26. https://0-doi-org.brum.beds.ac.uk/10.3390/scipharm89020026
Chicago/Turabian StyleJiso, Apisada, Laphatrada Yurasakpong, Sirorat Janta, Kulathida Chaithirayanon, and Anuchit Plubrukarn. 2021. "Exerting DNA Damaging Effects of the Ilimaquinones through the Active Hydroquinone Species" Scientia Pharmaceutica 89, no. 2: 26. https://0-doi-org.brum.beds.ac.uk/10.3390/scipharm89020026






