Multi-Omics Analysis to Generate Hypotheses for Mild Health Problems in Monkeys
Abstract
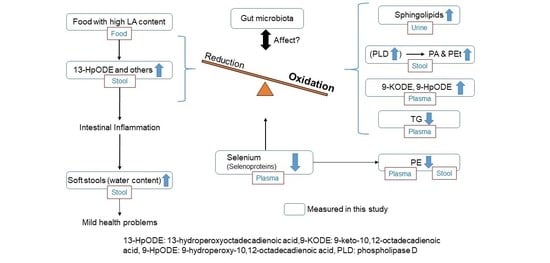
:1. Introduction
2. Results
2.1. Stool Water Content Analysis
2.2. Metabolomics
2.3. Fatty Acid Analysis
2.4. Lipidomics
2.5. Lipid Mediator Analysis
2.6. Metallomic Analysis
2.7. Microbiota Analysis
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Samples
4.3. Sample Preparation
4.4. Measuring Instruments and Measurement Parameters
4.5. Sequencing Data Processing and Analysis
4.6. Lipid Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Lipid Abbreviations
References
- Phillips, K.A.; Bales, K.L.; Capitanio, J.P.; Conley, A.; Czoty, P.W.; ’t Hart, B.A.; Hopkins, W.D.; Hu, S.-L.; Miller, L.A.; Nader, M.A.; et al. Why primate models matter. Am. J. Primatol. 2014, 76, 801–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colman, R.J. Non-human primates as a model for aging. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2733–2741. [Google Scholar] [CrossRef]
- Misra, B.B.; Bassey, E.; Bishop, A.C.; Kusel, D.T.; Cox, L.A.; Olivier, M. High-resolution gas chromatography/mass spectrometry metabolomics of non-human primate serum. Rapid Commun. Mass Spectrom. 2018, 32, 1497–1506. [Google Scholar] [CrossRef]
- Hird, D.W.; Anderson, J.H.; Bielitzki, J.T. Diarrhea in nonhuman primates: A survey of primate colonies for incidence rates and clinical opinion. Lab. Anim. Sci. 1984, 34, 465–470. [Google Scholar] [PubMed]
- Gottlieb, D.H.; Del Rosso, L.; Sheikhi, F.; Gottlieb, A.; McCowan, B.; Capitanio, J.P. Personality, environmental stressors, and diarrhea in Rhesus macaques: An interactionist perspective. Am. J. Primatol. 2018, 80, e22908. [Google Scholar] [CrossRef]
- Lovell, R.M.; Ford, A.C. Global Prevalence of and risk Factors for irritable bowel syndrome: A meta-analysis. Clin. Gastroenterol. Hepatol. 2012, 10, 712–721.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Liu, S.; Li, H.; Zhang, Z.; Zhang, Q.; Chen, L.; Zhao, Y.; Chen, Y.; Gu, J.; Min, L.; et al. Identification of gut microbiota and metabolites signature in patients with irritable bowel syndrome. Front. Cell. Infect. Microbiol. 2019, 9, 346. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Si, C.; Yu, Y.; Zhao, G.; Chen, L.; Zhao, Y.; Zhang, Z.; Li, H.; Chen, Y.; Min, L.; et al. Multi-omics Analysis of Gut Microbiota and Metabolites in Rats With Irritable Bowel Syndrome. Front. Cell. Infect. Microbiol. 2019, 9, 178. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.X.; Zhang, Y.; Qin, G.; Li, K.M.; Wei, W.; Li, S.Y.; Yao, S.K. Altered profiles of fecal metabolites correlate with visceral hypersensitivity and may contribute to symptom severity of diarrhea-predominant irritable bowel syndrome. World J. Gastroenterol. 2019, 25, 6416–6429. [Google Scholar] [CrossRef]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems biology and multi-omics integration: Viewpoints from the metabolomics research community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 2020, 14, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, S.M.; Sanghi, A.; Wu, S.; Snyder, M.P. A customizable analysis flow in integrative multi-omics. Biomolecules 2020, 10, 1606. [Google Scholar] [CrossRef] [PubMed]
- Wörheide, M.A.; Krumsiek, J.; Kastenmüller, G.; Arnold, M. Multi-omics integration in biomedical research—A metabolomics-centric review. Anal. Chim. Acta 2021, 1141, 144–162. [Google Scholar] [CrossRef]
- Bliss, D.Z.; Savik, K.; Jung, H.; Jensen, L.; LeMoine, M.; Lowry, A. Comparison of subjective classification of stool consistency and stool water content. J. Wound Ostomy Cont. Nurs. 1999, 26, 137–141. [Google Scholar]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Tito, R.Y.; Joossens, M.; Raes, J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016, 65, 57–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quell, J.D.; Römisch-Margl, W.; Colombo, M.; Krumsiek, J.; Evans, A.M.; Mohney, R.; Salomaa, V.; de Faire, U.; Groop, L.C.; Agakov, F.; et al. Automated pathway and reaction prediction facilitates in silico identification of unknown metabolites in human cohort studies. J. Chromatogr. B 2017, 1071, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Alvarez, J.A.; Kang, J.; Yu, T. Network marker selection for untargeted LC-MS metabolomics data. J. Proteome Res. 2017, 16, 1261–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basu, S.; Duren, W.; Evans, C.R.; Burant, C.F.; Michailidis, G.; Karnovsky, A. Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinformatics 2017, 33, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Guo, H.; Lu, Z.; Zhang, T.; Zhao, R.; Tao, N.; Wang, X.; Zhong, J. Reliability of LipidSearch software identification and its application to assess the effect of dry salting on the long-chain free fatty acid profile of tilapia muscles. Food Res. Int. 2020, 138, 109791. [Google Scholar] [CrossRef]
- Trzeciecka, A.; Stark, D.T.; Kwong, J.M.K.; Piqueras, M.; Bhattacharya, S.K.; Caprioli, J. Comparative lipid profiling dataset of the inflammation-induced optic nerve regeneration. Data Brief 2019, 24. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G.; De Kroon, A.I.P.M. Lipid map of the mammalian cell. J. Cell Sci. 2011, 124, 5–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Symons, J.L.; Cho, K.J.; Chang, J.T.; Du, G.; Waxham, M.N.; Hancock, J.F.; Levental, I.; Levental, K.R. Lipidomic atlas of mammalian cell membranes reveals hierarchical variation induced by culture conditions, subcellular membranes, and cell lineages. Soft Matter 2021, 17, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.F.; Savage, P.B.; Epand, R.M. Bacterial lipid composition and the antimicrobial efficacy of cationic steroid compounds (Ceragenins). Biochim. Biophys. Acta Biomembr. 2007, 1768, 2500–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2015, 40, 133–159. [Google Scholar] [CrossRef] [Green Version]
- Rohr, M.; Narasimhulu, C.A.; Keewan, E.; Hamid, S.; Parthasarathy, S. The dietary peroxidized lipid, 13-HPODE, promotes intestinal inflammation by mediating granzyme B secretion from natural killer cells. Food Funct. 2020, 11, 9526–9534. [Google Scholar] [CrossRef] [PubMed]
- Keewan, E.; Narasimhulu, C.A.; Rohr, M.; Hamid, S.; Parthasarathy, S. Are fried foods unhealthy? The dietary peroxidized fatty acid, 13-hpode, induces intestinal inflammation in vitro and in vivo. Antioxidants 2020, 9, 926. [Google Scholar] [CrossRef]
- Zoidis, E.; Seremelis, I.; Kontopoulos, N.; Danezis, G.P. Selenium-dependent antioxidant enzymes: Actions and properties of selenoproteins. Antioxidants 2018, 7, 66. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.; Lee, J.; Wu, C.; Guo, X.; Lee, B.J.; Chun, J.S.; Kim, J.H. The role of selenium metabolism and selenoproteins in cartilage homeostasis and arthropathies. Exp. Mol. Med. 2020, 52, 1198–1208. [Google Scholar] [CrossRef]
- Ingold, I.; Berndt, C.; Schmitt, S.; Doll, S.; Poschmann, G.; Buday, K.; Roveri, A.; Peng, X.; Freitas, F.P.; Seibt, T.; et al. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell 2018, 172, 409–422. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Suzuki, N.; Ogra, Y. Effect of gut microflora on nutritional availability of selenium. Food Chem. 2020, 319, 126537. [Google Scholar] [CrossRef]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The controversial role of human gut lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- Lee, G.; You, H.J.; Bajaj, J.S.; Joo, S.K.; Yu, J.; Park, S.; Kang, H.; Park, J.H.; Kim, J.H.; Lee, D.H.; et al. Distinct signatures of gut microbiome and metabolites associated with significant fibrosis in non-obese NAFLD. Nat. Commun. 2020, 11, 4982. [Google Scholar] [CrossRef]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019, 9, 287. [Google Scholar] [CrossRef]
- Watanabe, S.; Kameoka, S.; Shinozaki, N.O.; Kubo, R.; Nishida, A.; Kuriyama, M.; Takeda, A.K. A cross-sectional analysis from the Mykinso Cohort Study: Establishing reference ranges for Japanese gut microbial indices. Biosci. Microbiota Food Health 2021, 40, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Brunkwall, L.; Ericson, U.; Nilsson, P.M.; Orho-Melander, M.; Ohlsson, B. Self-reported bowel symptoms are associated with differences in overall gut microbiota composition and enrichment of Blautia in a population-based cohort. J. Gastroenterol. Hepatol. 2021, 36, 174–180. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 2015, 5, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakajima, H.; Takewaki, F.; Hashimoto, Y.; Kajiyama, S.; Majima, S.; Okada, H.; Senmaru, T.; Ushigome, E.; Nakanishi, N.; Hamaguchi, M.; et al. The effects of metformin on the gut microbiota of patients with type 2 diabetes: A two-center, quasi-experimental study. Life 2020, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Schoster, A.; Staempfli, H.R.; Guardabassi, L.G.; Jalali, M.; Weese, J.S. Comparison of the fecal bacterial microbiota of healthy and diarrheic foals at two and four weeks of life. BMC Vet. Res. 2017, 13, 144. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Huang, Y.; Lu, L.; Yang, W.; Huang, T.; Lin, Z.; Lin, C.; Kwan, H.; Wong, H.L.X.; Chen, Y.; et al. Saturated long-chain fatty acid-producing bacteria contribute to enhanced colonic motility in rats. Microbiome 2018, 6, 107. [Google Scholar] [CrossRef]
- Voshol, P.J.; Rensen, P.C.N.; van Dijk, K.W.; Romijn, J.A.; Havekes, L.M. Effect of plasma triglyceride metabolism on lipid storage in adipose tissue: Studies using genetically engineered mouse models. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2009, 1791, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Ohtsu, H.; Yakabe, Y.; Yamazaki, M.; Murakami, H.; Abe, H. Plasma lipid profiles and redox status are modulated in a ketogenic diet-induced chicken model of ketosis. J. Poult. Sci. 2013, 50, 212–218. [Google Scholar] [CrossRef] [Green Version]
- Benjamin, B.; Wada, Y.; Grundy, S.M.; Szuszkiewicz-Garcia, M.; Vega, G.L. Fatty acid oxidation in normotriglyceridemic men. J. Clin. Lipidol. 2016, 10, 283–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Hoffmann, F.W.; Marciel, M.P.; Page, K.E.; Williams-Aduja, M.A.; Akana, E.N.L.; Gojanovich, G.S.; Gerschenson, M.; Urschitz, J.; Moisyadi, S.; et al. Upregulated ethanolamine phospholipid synthesis via selenoprotein I is required for effective metabolic reprogramming during T cell activation. Mol. Metab. 2021, 47, 101170. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.; Parinandi, N.; Wang, L.; Usatyuk, P.; Natarajan, V. Phospholipase D/phosphatidic acid signal transduction: Role and physiological significance in lung. Mol. Cell. Biochem. 2002, 234–235, 99–109. [Google Scholar] [CrossRef]
- Tappia, P.S.; Dent, M.R.; Dhalla, N.S. Oxidative stress and redox regulation of phospholipase D in myocardial disease. Free Radic. Biol. Med. 2006, 41, 349–361. [Google Scholar] [CrossRef]
- Ramrakhiani, L.; Chand, S. Recent progress on phospholipases: Different sources, assay methods, industrial potential and pathogenicity. Appl. Biochem. Biotechnol. 2011, 164, 991–1022. [Google Scholar] [CrossRef]
- Isaksson, A.; Walther, L.; Hansson, T.; Andersson, A.; Alling, C. Phosphatidylethanol in blood (B-PEth): A marker for alcohol use and abuse. Drug Test. Anal. 2011, 3, 195–200. [Google Scholar] [CrossRef]
- Liu, J.J.; Ghosh, S.; Kovalik, J.P.; Ching, J.; Choi, H.W.; Tavintharan, S.; Ong, C.N.; Sum, C.F.; Summers, S.A.; Tai, E.S.; et al. Profiling of plasma metabolites suggests altered mitochondrial fuel usage and remodeling of sphingolipid metabolism in individuals with Type 2 diabetes and kidney disease. Kidney Int. Rep. 2017, 2, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Bhat, O.M.; Yuan, X.; Li, G.; Lee, R.; Li, P.L. Sphingolipids and redox signaling in renal regulation and chronic kidney diseases. Antioxid. Redox Signal. 2018, 28, 1008–1026. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Zhang, X.; Lu, Y.; Chen, H. New insights in intestinal oxidative stress damage and the health intervention effects of nutrients: A review. J. Funct. Foods 2020, 75, 104248. [Google Scholar] [CrossRef]
- Vona, R.; Pallotta, L.; Cappelletti, M.; Severi, C.; Matarrese, P. The impact of oxidative stress in human pathology: Focus on gastrointestinal disorders. Antioxidants 2021, 10, 201. [Google Scholar] [CrossRef]
- Knuplez, E.; Marsche, G. An updated review of pro-and anti-inflammatory properties of plasma lysophosphatidylcholines in the vascular system. Int. J. Mol. Sci. 2020, 21, 4501. [Google Scholar] [CrossRef] [PubMed]
- Kita, Y.; Takahashi, T.; Uozumi, N.; Shimizu, T. A multiplex quantitation method for eicosanoids and platelet-activating factor using column-switching reversed-phase liquid chromatography-tandem mass spectrometry. Anal. Biochem. 2005, 342, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A.; Mcnally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Yamada, M.; Kita, Y.; Kohira, T.; Yoshida, K.; Hamano, F.; Tokuoka, S.M.; Shimizu, T. A comprehensive quantification method for eicosanoids and related compounds by using liquid chromatography/mass spectrometry with high speed continuous ionization polarity switching. J. Chromatogr. B 2015, 995–996, 74–84. [Google Scholar] [CrossRef]
- Tácito, L.H.B.; Yamada, L.N.; de Pinhel, M.A.S.; Yugar-Toledo, J.C.; Souza, D.R.S. Influence of Apolipoprotein E on the lipid profile and postprandial triglyceride levels in Brazilian postmenopausal women with artery disease. Clin. Med. Insights Cardiol. 2017, 11. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]









| Monkey | Water % | Age (Years) |
|---|---|---|
| 1 | 57.93 | 5.8 |
| 2 * | 74.50 | 5.1 |
| 3 * | 74.07 | 4.2 |
| 4 | 65.52 | 4.7 |
| 5 * | 77.21 | 5.0 |
| 6 | 71.46 | 5.1 |
| 7 * | 79.45 | 6.3 |
| 8 | 72.62 | 5.1 |
| 9 | 67.80 | 5.6 |
| 10 | 69.09 | 6.3 |
| 11 | 72.98 | 6.2 |
| 12 | 75.36 | 5.1 |
| 13 | 62.24 | 6.3 |
| 14 | 57.89 | 5.3 |
| 15 | 66.60 | 6.1 |
| 16 | 64.70 | 4.2 |
| 17 | 72.67 | 3.9 |
| 18 | 67.69 | 4.4 |
| 19 | 65.50 | 3.8 |
| 20 * | 76.92 | 4.0 |
| Lipid Name | Correlation Coefficient | p-Value |
|---|---|---|
| 13-HpODE | 0.732 | 0.00024 |
| 9-HpODE | 0.717 | 0.00038 |
| 12,13-EpOME | 0.71 | 0.00045 |
| 13-HOTrE | 0.659 | 0.0016 |
| 13-HODE | 0.651 | 0.0019 |
| 13-KODE | 0.618 | 0.0037 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamano, F.; Tokuoka, S.M.; Ishibashi, M.; Yokoi, Y.; Tourlousse, D.M.; Kita, Y.; Sekiguchi, Y.; Yasui, H.; Shimizu, T.; Oda, Y. Multi-Omics Analysis to Generate Hypotheses for Mild Health Problems in Monkeys. Metabolites 2021, 11, 701. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11100701
Hamano F, Tokuoka SM, Ishibashi M, Yokoi Y, Tourlousse DM, Kita Y, Sekiguchi Y, Yasui H, Shimizu T, Oda Y. Multi-Omics Analysis to Generate Hypotheses for Mild Health Problems in Monkeys. Metabolites. 2021; 11(10):701. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11100701
Chicago/Turabian StyleHamano, Fumie, Suzumi M. Tokuoka, Megumi Ishibashi, Yasuto Yokoi, Dieter M. Tourlousse, Yoshihiro Kita, Yuji Sekiguchi, Hiroyuki Yasui, Takao Shimizu, and Yoshiya Oda. 2021. "Multi-Omics Analysis to Generate Hypotheses for Mild Health Problems in Monkeys" Metabolites 11, no. 10: 701. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11100701