Analyzing the Metabolomic Profile of Yellowtail (Seriola quinquerdiata) by Capillary Electrophoresis–Time of Flight Mass Spectrometry to Determine Geographical Origin
Abstract
:1. Introduction
2. Results and Discussion
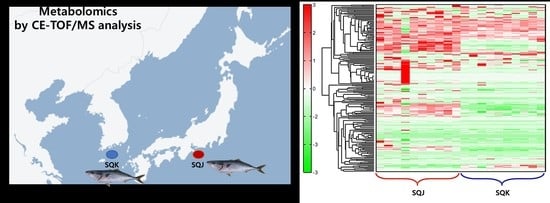
2.1. Metabolomics Profile
2.2. Differences in Organic Acid Metabolites in the Tricarboxylic Acid (TCA) Cycle
2.3. Differences in Metabolites in Amino Acid Metabolism
2.4. Differences in Metabolites in Other Quantified Compounds
3. Materials and Methods
3.1. Sample Collection and Preparation
3.2. Metabolite Extraction
3.3. CE-TOF/MS Analysis
3.4. Metabolomics Profile Analysis
3.5. Statistics Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, C.M.; Lee, M.N.; Kim, E.M.; Park, J.Y.; Kim, G.D.; Noh, J.K. Development and Genetic Diversity Analysis of Microsatellite Markers Using Next-generation Sequencing in Seriola quinqueradiata. J. Life Sci. 2020, 30, 291–297. [Google Scholar]
- Kim, S.W.; Ahn, J.S.; Lee, Y. The Effect of Interannual Variations in Water Temperature on the Yellowtail Catch, Seriola Quinqueradiata, in the Eastern Part of the Korean Peninsula. J. Korean Soc. Mar. Environ. Saf. 2017, 23, 909–917. [Google Scholar] [CrossRef]
- Ministry of Oceans and Fisheries. Statistics System of Korean Ministry of Oceans and Fisheries. Available online: https://www.mof.go.kr/statPortal/main/portalMain.do (accessed on 15 August 2021).
- Choi, J.Y.; Bang, K.H.; Han, K.Y.; Noh, B.S. Discrimination analysis of the geographical origin of foods. Korean J. Food Sci. Technol. 2012, 44, 503–525. [Google Scholar] [CrossRef]
- Jung, Y.; Lee, J.; Kwon, J.; Lee, K.S.; Ryu, D.H.; Hwang, G.S. Discrimination of the geographical origin of beef by 1H NMR-based metabolomics. J. Agric. Food Chem. 2010, 58, 10458–10466. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Guo, B.; Wei, Y.; Fan, M. Multi-Element analysis for determining the geographical origin of mutton from different regions of China. Food Chem. 2011, 124, 1151–1156. [Google Scholar] [CrossRef]
- Kang, D.J.; Lee, S.K.; Jin, D.H.; Choi, S.J. Random amplified polymorphic DNA analysis for origin identification of olive flounder (Paralichthys olivaceus) and redlip croaker (Pseudosciaena polyactis). J. Life Sci. 2006, 16, 88–94. [Google Scholar]
- Song, I.S.; Jin, D.H.; Choi, S.J.; Lee, S.G. Polymorphism analysis of the ND-4 gene for the origin determination of olive flounder, Paralichthys olivaceus. J. Life Sci. 2004, 16, 627–635. [Google Scholar]
- Brown, M.; Dunn, W.B.; Dobson, P.; Patel, Y.; Winder, C.L.; Francis-McIntyre, S.; Begley, P.; Carroll, K.; Broadhurst, D.; Tseng, A.; et al. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst 2009, 134, 1322–1332. [Google Scholar] [CrossRef]
- Dhanasekaran, A.R.; Pearson, J.L.; Ganesan, B.; Weimer, B.C. Metabolome searcher: A high throughput tool for metabolite identification and metabolic pathway mapping directly from mass spectrometry and using genome restriction. BMC Bioinform. 2015, 16, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ghisoni, S.; Lucini, L.; Angilletta, F.; Rocchetti, G.; Farinelli, D.; Tombesi, S.; Trevisan, M. Discrimination of extra-virgin-olive oils from different cultivars and geographical origins by untargeted metabolomics. Food Res. Int. 2019, 121, 746–753. [Google Scholar] [CrossRef]
- Lee, B.M.; Lee, E.M.; Kang, D.J.; Seo, J.A.; Choi, H.K.; Kim, Y.S.; Lee, D.Y. Discovery study of integrative metabolic profiles of sesame seeds cultivated in different countries. LWT 2020, 129, 109454. [Google Scholar] [CrossRef]
- Zhao, G.; Zhao, W.; Han, L.; Ding, J.; Chang, Y. Metabolomics analysis of sea cucumber (Apostichopus japonicus) in different geographical origins using UPLC–Q-TOF/MS. Food Chem. 2020, 333, 127453. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, J.; Zhao, C.; Zhou, H.; Li, Y.; Zhang, J.; Li, L.; Hu, C.; Li, W.; Peng, X.; et al. A metabolomics study delineating geographical location-associated primary metabolic changes in the leaves of growing tobacco plants by GC-MS and CE-MS. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef]
- Bodner, G.M. Metabolism part II: The tricarboxylic acid (TCA), citric acid, or Krebs cycle. J. Chem. Educ. 1986, 63, 673. [Google Scholar] [CrossRef]
- Shambaugh, G.E., III. Urea biosynthesis I. The urea cycle and relationships to the citric acid cycle. Am. J. Clin. Nutr. 1977, 30, 2083–2087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berka, R. The Transport of Live Fish: A Review; Food and Agriculture Organization of the United Nations: Rome, Italy, 1986; Volume 48, pp. 1–3. [Google Scholar]
- Hertz, Y.; Madar, Z.; Hepher, B.; Gertler, A. Glucose metabolism in the common carp (Cyprinus carpio L.): The effects of cobalt and chromium. Aquaculture 1989, 76, 255–267. [Google Scholar] [CrossRef]
- Nakada, M.K. Yellowtail culture development and solutions for the future. Rev. Fish. Sci. 2002, 10, 559–575. [Google Scholar] [CrossRef]
- Nakada, M. Capture-Based aquaculture of yellowtail. Capture-Based aquaculture. Global overview. FAO Fish. Tech. Pap. 2008, 508, 199–215. [Google Scholar]
- Chang, D.S.; Yoo, J.T.; Kim, B.Y.; Lee, S.J.; Kwon, D.H.; Koo, J.H.; Ahn, G.M.; Oh, I.Y. A characteristics on the forming of fishing ground and population ecological study of Yellow tail, Seriola quinqueradiata, in the coastal waters off Gim-nyeong of Jeju Island, Korea. J. Korean Soc. Fish. Ocean. Technol. 2010, 46, 406–415. [Google Scholar] [CrossRef] [Green Version]
- Craig, S.; Helfrich, L.A.; Kuhn, D.; Schwarz, M.H. Understanding fish nutrition, feeds, and feeding. Va. Coop. Ext. 2017, 420–256, 1–6. [Google Scholar]
- Jang, M.S.; Kang, Y.J.; Kim, K.W.; Kim, K.D.; Lee, H.Y.; Heo, S.B. Quality characteristics of cultured olive flounder Paralichthys olivaceus fed with extruded pellets; I. Comparison of fatty acid and amino acid contents. Korean J. Food Sci. Technol. 2009, 41, 42–49. [Google Scholar]
- Walton, M.J.; Cowey, C.B.; Adron, J.W. Methionine metabolism in rainbow trout fed diets of differing methionine and cystine content. J. Nutr. 1982, 112, 1525–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, A.M., Jr. Nutrition, digestion, and energy utilization. Fish. Physiol. 1969, 1, 391–432. [Google Scholar]
- Blaxter, K.L.; Wainman, F.W. The fasting metabolism of cattle. Br. J. Nutr. 1966, 20, 103–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, G.M.; Ahn, Y.S.; Shin, D.M.; Kim, H.S.; Kim, H.J.; Yoon, M.S.; Heo, M.S.; Kim, J.S. Comparison of muscle color, taste and nutrition components between red seabreams cultured by feeding and starving. J. Korean Soc. Food Sci. Nutr. 2008, 37, 1142–1147. [Google Scholar] [CrossRef]
- Knox, D.; Walton, M.J.; Cowey, C.B. Distribution of enzymes of glycolysis and gluconeogenesis in fish tissues. Mar. Biol. 1980, 56, 7–10. [Google Scholar] [CrossRef]
- Moose, P.R. An investigation of gluconeogenesis in marine teleosts, and the effect of long-term exercise on hepatic gluconeogenesis. Comp. Biochem. Physiol. Part. B Comp. Biochem. 1980, 67, 583–592. [Google Scholar] [CrossRef]
- Sargent, J.R.; Tocher, D.R.; Bell, J.G. The Lipids. In Fish Nutrition, 3rd ed.; Hardy, R., Ed.; Elsevier (Academic Press): San Diego, CA, USA, 2002; pp. 181–257. [Google Scholar]
- Jürss, K.; Bastrop, R. Amino acid metabolism in fish. Biochem. Mol. Biol. Fishes 1995, 4, 159–189. [Google Scholar]
- Yokoyama, H.; Tadokoro, D.; Miura, M. Quantification of waste feed and fish faeces in sediments beneath yellowtail pens and possibility to reduce waste loading by co-culturing with sea cucumbers: An isotopic study. Aquac. Res. 2015, 46, 918–927. [Google Scholar] [CrossRef]
- Das, G.; Patra, J.K.; Lee, S.Y.; Kim, C.; Park, J.G.; Baek, K.H. Analysis of metabolomic profile of fermented Orostachys japonicus A. Berger by capillary electrophoresis time of flight mass spectrometry. PLoS ONE 2017, 12, e0181280. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Concentration (nmol·g−1) | Comparative Analysis | |||
|---|---|---|---|---|
| SQK | SQJ | Ratio 2 | p-Value 3 | |
| Guanosine | 10 ± 2 1 | 22 ± 6.7 | 2.2 | 0.00023 *** |
| Hypoxanthine | 55 ± 22 | 120 ± 79 | 2.2 | 0.031 * |
| Guanine | 1.0 ± 0.2 | 2.9 ± 1.6 | 2.8 | 0.006 ** |
| Sarcosine | 3.5 ± 1.2 | 13 ± 4.0 | 3.6 | 0.000028 *** |
| SQK | SQJ | |||
|---|---|---|---|---|
| Sample Number | Full Length (cm) | Weight (kg) | Full Length (cm) | Weight (kg) |
| 1 | 84.0 | 4.92 | 74.0 | 4.46 |
| 2 | 66.5 | 3.95 | 78.2 | 4.94 |
| 3 | 71.1 | 3.99 | 75.0 | 4.85 |
| 4 | 72.3 | 4.15 | 75.6 | 4.10 |
| 5 | 71.9 | 4.12 | 79.0 | 5.50 |
| 6 | 70.9 | 3.93 | 80.8 | 5.40 |
| 7 | 71.8 | 4.14 | 77.0 | 4.97 |
| 8 | 68.0 | 3.86 | 76.4 | 4.58 |
| 9 | 72.6 | 4.06 | 77.0 | 3.98 |
| 10 | 72.7 | 4.56 | 73.8 | 4.07 |
| Mean ± S.D. | 72.7 ± 4.63 | 4.2 ± 0.33 | 76.7 ± 2.22 | 4.7 ± 0.54 |
| Cationic Metabolites (Cation Mode) | Anionic Metabolites (Anion Mode) | |
|---|---|---|
| Capillary | fused silica capillary of 50 μm × 80 cm | |
| Run buffer | Cation Buffer Soln. (p/n: H3301-1001) | Anion Buffer Soln. (p/n: I3302-1023) |
| Rinse buffer | Cation Buffer Soln. (p/n: H3301-1001) | Anion buffer Soln. (p/n: I3302-1023) |
| Sample injection | Pressure injection 55 mbar, 10 s | Pressure injection 50 mbar, 22 s |
| CE voltage | Positive, 30 kV | Positive, 30 kV |
| MS ionization | Electrospray ionization (ESI) Positive | ESI negative |
| MS Capillary voltage | 4000 V | 3500 V |
| MS scan range | m/z 50–1000 | m/z 50–1000 |
| Sheath liquid | HMT Sheath Liquid (p/n: H3301-1021) | HMT Sheath Liquid (p/n: H3301-1021) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.; Yang, J.; Cha, E.; Kim, H.; Lee, Y.; Kim, S.; Choi, I.; Yang, J. Analyzing the Metabolomic Profile of Yellowtail (Seriola quinquerdiata) by Capillary Electrophoresis–Time of Flight Mass Spectrometry to Determine Geographical Origin. Metabolites 2021, 11, 793. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11110793
Shin J, Yang J, Cha E, Kim H, Lee Y, Kim S, Choi I, Yang J. Analyzing the Metabolomic Profile of Yellowtail (Seriola quinquerdiata) by Capillary Electrophoresis–Time of Flight Mass Spectrometry to Determine Geographical Origin. Metabolites. 2021; 11(11):793. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11110793
Chicago/Turabian StyleShin, Jiyoung, Junho Yang, Eunji Cha, Hyunsuk Kim, Yoonhyeung Lee, Soi Kim, Iseul Choi, and Jiyoung Yang. 2021. "Analyzing the Metabolomic Profile of Yellowtail (Seriola quinquerdiata) by Capillary Electrophoresis–Time of Flight Mass Spectrometry to Determine Geographical Origin" Metabolites 11, no. 11: 793. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11110793