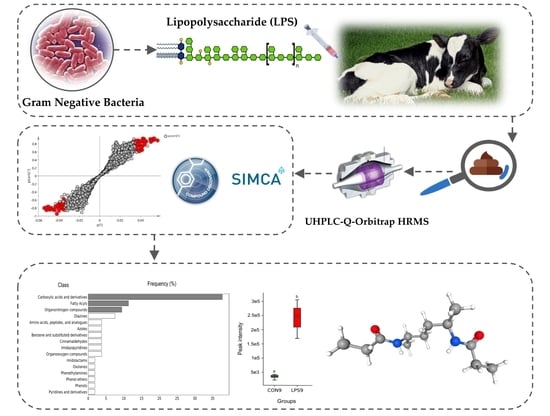
Metabolic Fingerprinting of Feces from Calves, Subjected to Gram-Negative Bacterial Endotoxin
Abstract
:1. Introduction
2. Results
2.1. Acute Phase Response
2.2. Metabolic Fingerprinting by UHPLC-HRMS
2.3. Identification of Potential UHPLC-HRMS Metabolite Markers
2.4. Identification of Potential Pathway Alterations
3. Discussion
4. Materials and Methods
4.1. Calves, Treatments and Study Design
4.2. Acute Phase Response Evaluation and Sample Collection
4.3. UHPLC-Q-Orbitrap-HRMS Based Metabolomic Evaluation
4.4. Data Analysis
4.5. Identification of Potential APR Metabolite Markers in Calves
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortiz-Pelaez, A.; Pritchard, D.; Pfeiffer, D.; Jones, E.; Honeyman, P.; Mawdsley, J. Calf mortality as a welfare indicator on British cattle farms. Vet. J. 2008, 176, 177–181. [Google Scholar] [CrossRef]
- Michaels, F.; Banks, K. Contribution of various host factors to resistance to experimentally induced bacterial endotoxemia in calves. Am. J. Vet. Res. 1988, 49, 557–562. [Google Scholar]
- Pardon, B.; De Bleecker, K.; Hostens, M.; Callens, J.; Dewulf, J.; Deprez, P. Longitudinal study on morbidity and mortality in white veal calves in Belgium. BMC Vet. Res. 2012, 8, 26. [Google Scholar] [CrossRef] [Green Version]
- Fecteau, G.; Smith, B.P.; George, L.W. Septicemia and meningitis in the newborn calf. Vet. Clin. N. Am. Food Anim. Pract. 2009, 25, 195–208. [Google Scholar] [CrossRef]
- Plessers, E.; Watteyn, A.; Wyns, H.; Pardon, B.; De Backer, P.; Croubels, S. Study of the immunomodulatory properties of gamithromycin and dexamethasone in a lipopolysaccharide inflammation model in calves. Res. Vet. Sci. 2015, 103, 218–223. [Google Scholar] [CrossRef]
- Smith, G. Weakness and/or depressed mentation. Res. Vet. Sci. 2015, 105, 218–223. [Google Scholar]
- Basoglu, A.; Sen, I.; Meoni, G.; Tenori, L.; Naseri, A. NMR-based plasma metabolomics at set intervals in newborn dairy calves with severe sepsis. Mediat. Inflamma. 2018, 2018, 8016510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suhre, K.; Gieger, C. Genetic variation in metabolic phenotypes: Study designs and applications. Nat. Rev. Genet. 2012, 13, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant. Mol. Biol. 2002, 155–171. [Google Scholar] [CrossRef]
- Claus, S.P.; Swann, J.R. Nutrimetabonomics: Applications for nutritional sciences, with specific reference to gut microbial interactions. Annu. Rev. Food Sci. Technol. 2013, 4, 381–399. [Google Scholar] [CrossRef]
- De Paepe, E.; Van Meulebroek, L.; Rombouts, C.; Huysman, S.; Verplanken, K.; Lapauw, B.; Wauters, J.; Hemeryck, L.Y.; Vanhaecke, L. A validated multi-matrix platform for metabolomic fingerprinting of human urine, feces and plasma using ultra-high performance liquid-chromatography coupled to hybrid orbitrap high-resolution mass spectrometry. Anal. Chim. Acta 2018, 1033, 108–118. [Google Scholar] [CrossRef]
- Laíns, I.; Gantner, M.; Murinello, S.; Lasky-Su, J.A.; Miller, J.W.; Friedlander, M.; Husain, D. Metabolomics in the study of retinal health and disease. Prog. Retin. Eye Res. 2019, 69, 57–79. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.M.; Darzi, A.W.; Takats, Z.; Lindon, J.C. Metabolic phenotyping in clinical and surgical environments. Nature 2012, 491, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Van Meulebroek, L.; Cameron, S.; Plekhova, V.; De Spiegeleer, M.; Wijnant, K.; Michels, N.; De Henauw, S.; Lapauw, B.; Takats, Z.; Vanhaecke, L. Rapid LA-REIMS and comprehensive UHPLC-HRMS for metabolic phenotyping of feces. Talanta 2020, 217, 121043. [Google Scholar] [CrossRef] [PubMed]
- Basoglu, A.; Baspinar, N.; Tenori, L.; Hu, X.; Yildiz, R. NMR based metabolomics evaluation in neonatal calves with acute diarrhea and suspected sepsis: A new approach for biomarker/S. Metabolomics 2014, 4, 1. [Google Scholar]
- Basoglu, A.; Baspinar, N.; Tenori, L.; Vignoli, A.; Yildiz, R. Plasma metabolomics in calves with acute bronchopneumonia. Metabolomics 2016, 12, 128. [Google Scholar] [CrossRef]
- Gray, D.W.; Welsh, M.D.; Doherty, S.; Mansoor, F.; Chevallier, O.P.; Elliott, C.T.; Mooney, M.H. Identification of systemic immune response markers through metabolomic profiling of plasma from calves given an intra-nasally delivered respiratory vaccine. Vet. Res. 2015, 46, 7. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.Z.; Cui, D.A.; Wu, X.H.; Hui, W.; Yan, Z.T.; Ding, X.Z.; Wang, S.Y. Serum Metabolomics Revealed the Differential Metabolic Pathway in Calves with Severe Clinical Diarrhea Symptoms. Animals 2020, 10, 769. [Google Scholar] [CrossRef]
- Plessers, E.; Wyns, H.; Watteyn, A.; Pardon, B.; De Backer, P.; Croubels, S. Characterization of an intravenous lipopolysaccharide inflammation model in calves with respect to the acute-phase response. Vet. Immunol. Immunopathol. 2015, 163, 46–56. [Google Scholar] [CrossRef]
- Hill, T.M.; Vandehaar, M.J.; Sordillo, L.M.; Catherman, D.R.; Bateman, H.G., 2nd; Schlotterbeck, R.L. Fatty acid intake alters growth and immunity in milk-fed calves. J. Dairy Sci. 2011, 94, 3936–3948. [Google Scholar] [CrossRef] [Green Version]
- Karcher, E.L.; Hill, T.M.; Bateman, H.G., 2nd; Schlotterbeck, R.L.; Vito, N.; Sordillo, L.M.; Vandehaar, M.J. Comparison of supplementation of n-3 fatty acids from fish and flax oil on cytokine gene expression and growth of milk-fed Holstein calves. J. Dairy Sci. 2014, 97, 2329–2337. [Google Scholar] [CrossRef] [Green Version]
- Bible, M.R. Influence of Curcumin on Growth Performance and Immune Status of Nursery Pigs. Ph.D. Thesis, Oklahoma State University, Oklahoma, OK, USA, 2013. [Google Scholar]
- Vanden Bussche, J.; Marzorati, M.; Laukens, D.; Vanhaecke, L. Validated High Resolution Mass Spectrometry-Based Approach for Metabolomic Fingerprinting of the Human Gut Phenotype. Anal. Chem. 2015, 87, 10927–10934. [Google Scholar] [CrossRef]
- Kamel Oroumieh, S.; Vanhaecke, L.; Valizadeh, R.; Van Meulebroek, L.; Naserian, A.A. Effect of nanocurcumin and fish oil as natural anti-inflammatory compounds vs. glucocorticoids in a lipopolysaccharide inflammation model on Holstein calves’ health status. Heliyon 2021, 7, e058940. [Google Scholar] [CrossRef]
- Gao, J.; Tarcea, V.G.; Karnovsky, A.; Mirel, B.R.; Weymouth, T.E.; Beecher, C.W.; Cavalcoli, J.D.; Athey, B.D.; Omenn, G.S.; Burant, C.F.; et al. Metscape: A Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics 2010, 26, 971–973. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Badea, G.I.; Radu, G.L. Introductory Chapter: Carboxylic Acids-Key Role in Life Sciences. In Carboxylic Acid: Key Role in Life Sciences; IntechOpen: London, UK, 2018. [Google Scholar]
- Chalmers, R. Organic Acids in Man: Analytical Chemistry, Biochemistry and Diagnosis of the Organic Acidurias; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Nakamura, T.; Tabeke, K.; Terada, A.; Kudoh, K.; Yamada, N.; Arai, Y.; Kikuchi, H. Short-chain carboxylic acid in the feces in patients with pancreatic insufficiency. Acta Gastroenterol. Belg. 1993, 56, 326–331. [Google Scholar]
- Niederman, R.; Zhang, J.; Kashket, S. Short-chain carboxylic-acid-stimulated, PMN-mediated gingival inflammation. Crit. Rev. Oral. Biol. Med. 1997, 8, 269–290. [Google Scholar] [CrossRef]
- D’Acquisto, F.; Carnuccio, R.; d’Ischia, M.; Misuraca, G. 5,6-Dihydroxyindole-2-carboxylic acid, a diffusible melanin precursor, is a potent stimulator of lipopolysaccharide-induced production of nitric oxide by J774 macrophages. Life Sci. 1995, 57, PL401–PL406. [Google Scholar] [CrossRef]
- Mitrokhin, S.D.; Ivanov, A.A. The significance of individual carboxylic acids in the feces in the diagnosis of dysbacteriosis. Zh. Mikrobiol. Epidemiol. Immunobiol. 1995, 4, 99–101. [Google Scholar]
- Huang, P.; Liu, Y. A Reasonable Diet Promotes Balance of Intestinal Microbiota: Prevention of Precolorectal Cancer. Biomed. Res. Int. 2019, 2019, 3405278. [Google Scholar] [CrossRef]
- Dong, X.; Feng, X.; Liu, J.; Xu, Y.; Pan, Q.; Ling, Z.; Yu, J.; Yang, J.; Li, L.; Cao, H. Characteristics of Intestinal Microecology during Mesenchymal Stem Cell-Based Therapy for Mouse Acute Liver Injury. Stem Cells Int. 2019, 2019, 2403793. [Google Scholar] [CrossRef] [Green Version]
- Feunang, Y.D.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [Green Version]
- Divito, E.B.; Cascio, M. Metabolism, physiology, and analyses of primary fatty acid amides. Chem. Rev. 2013, 113, 7343–7353. [Google Scholar] [CrossRef]
- Pillarisetti, S.; Alexander, C.W.; Khanna, I. Pain and beyond: Fatty acid amides and fatty acid amide hydrolase inhibitors in cardiovascular and metabolic diseases. Drug Discov. Today 2009, 14, 1098–1111. [Google Scholar] [CrossRef] [PubMed]
- Maccarrone, M.; De Petrocellis, L.; Bari, M.; Fezza, F.; Salvati, S.; Di Marzo, V.; Finazzi-Agro, A. Lipopolysaccharide downregulates fatty acid amide hydrolase expression and increases anandamide levels in human peripheral lymphocytes. Arch. Biochem. Biophys. 2001, 393, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.; Gottfried-Blackmore, A.; Milner, T.A.; McEwen, B.S.; Bulloch, K. Steroid hormone receptor expression and function in microglia. Glia 2008, 56, 659–674. [Google Scholar] [CrossRef] [PubMed]
- Im, H.-J.; Park, N.-H.; Kwon, Y.-J.; Shin, S.; Kim, D.; Chun, Y.-J. Bacterial lipopolysaccharides induce steroid sulfatase expression and cell migration through IL-6 pathway in human prostate cancer cells. Biomol. Ther. 2012, 20, 556. [Google Scholar] [CrossRef] [Green Version]
- Rhen, T.; Cidlowski, J.A. Antiinflammatory action of glucocorticoids—New mechanisms for old drugs. N. Engl. J. Med. 2005, 353, 1711–1723. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, D.C.; da Silva Lima, F.; Sartori, T.; Santos, A.C.A.; Rogero, M.M.; Fock, R.A. Glutamine metabolism and its effects on immune response: Molecular mechanism and gene expression. Nutrire 2016, 41, 14. [Google Scholar] [CrossRef] [Green Version]
- Boutry, C.; Matsumoto, H.; Bos, C.; Moinard, C.; Cynober, L.; Yin, Y.; Tomé, D.; Blachier, F. Decreased glutamate, glutamine and citrulline concentrations in plasma and muscle in endotoxemia cannot be reversed by glutamate or glutamine supplementation: A primary intestinal defect? Amino Acids 2012, 43, 1485–1498. [Google Scholar] [CrossRef]
- Nielsen, S.S.; Grofte, T.; Tygstrup, N.; Vilstrup, H. Effect of lipopolysaccharide on in vivo and genetic regulation of rat urea synthesis. Liver Int. 2005, 25, 177–183. [Google Scholar] [CrossRef]
- Abad, B.; Mesonero, J.E.; Salvador, M.T.; Garcia-Herrera, J.; Rodriguez-Yoldi, M.J. Effect of lipopolysaccharide on small intestinal L-leucine transport in rabbit. Dig. Dis. Sci. 2001, 46, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Zaynagetdinov, R.; Yull, F.E.; Polosukhin, V.V.; Gleaves, L.A.; Tanjore, H.; Young, L.R.; Peterson, T.E.; Manning, H.C.; Prince, L.S. Molecular imaging of folate receptor β–positive macrophages during acute lung inflammation. Am. J. Respir. Cell Mol. Biol. 2015, 53, 50–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glynn, S.A.; Albanes, D. Folate and cancer: A review of the literature. Nutr. Cancer 1994, 22, 101–119. [Google Scholar] [CrossRef]
- Council, N.R. Nutrient Requirements of Dairy Cattle; National Academies Press: Washington, DC, USA, 2001. [Google Scholar]
- Kamleh, M.A.; Ebbels, T.M.; Spagou, K.; Masson, P.; Want, E.J. Optimizing the use of quality control samples for signal drift correction in large-scale urine metabolic profiling studies. Anal. Chem. 2012, 84, 2670–2677. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [Green Version]







| In-House Identifier (ID) | m/z | RT (min) | Chemical Formula | Adduct Ion | VIP 1 | p-Value 2 | Average Fold-Change 3 |
|---|---|---|---|---|---|---|---|
| 1 | 88.11257 | 3.35 | C5H13N | [M+H]+ | 3.02 | 0.00321 | 33.14 |
| 2 | 114.09153 | 1.11 | C6H11NO | [M+H]+ | 2.07 | 0.02781 | 0.19 |
| 3 | 133.049116 | 1.94 | C3H8N3O3 | [M-H]− | 1.95 | 0.04554 | 0.11 |
| 4 | 137.05927 | 7.65 | C6H8N3O | [M-H]− | 2.78 | 0.01925 | 0.02 |
| 5 | 146.11743 | 6.22 | C7H15NO2 | [M+H]+ | 2.42 | 0.02418 | 11.73 |
| 6 | 147.06314 | 9.31 | C4H8N3O3 | [M+H]+ | 1.97 | 0.02539 | 0.21 |
| 7 | 155.04495 | 4.13 | C6H6N2O3 | [M+H]+ | 2.09 | 0.03568 | 2.84 |
| 8 | 155.04480 | 1.86 | C4H6N5O2 | [M-H]− | 2.11 | 0.02501 | 0.12 |
| 9 | 157.06067 | 1.85 | C6H8N2O3 | [M+H]+ | 2.18 | 0.00486 | 0.11 |
| 10 | 156.06519 | 4.74 | C5H9N4O2 | [M-H]− | 2.14 | 0.00805 | 20.28 |
| 11 | 160.13307 | 7.47 | C8H17NO2 | [M+H]+ | 1.92 | 0.00807 | 5.90 |
| 12 | 165.05450 | 7.64 | C9H8O3 | [M+H]+ | 2.89 | 0.02354 | 0.02 |
| 13 | 165.02924 | 6.65 | C5H4N5O2 | [M-H]− | 2.00 | 0.04593 | 0.17 |
| 14 | 179.09273 | 2.21 | C8H10N4O | [M+H]+ | 2.34 | 0.02721 | 0.06 |
| 15 | 180.08516 | 2.21 | C4H11N4O4 | [M+H]+ | 2.02 | 0.04577 | 0.10 |
| 16 | 181.04941 | 7.65 | C7H8N3O3 | [M-H]− | 2.69 | 0.02264 | 0.03 |
| 17 | 196.14429 | 6.77 | C10H17N3O | [M+H]+ | 2.21 | 0.01398 | 15.15 |
| 18 | 205.13675 | 5.74 | C9H20N2OS | [M+H]+ | 2.69 | 0.00040 | 20.51 |
| 19 | 215.17520 | 8.87 | C11H22N2O2 | [M+H]+ | 2.67 | 0.00220 | 18.92 |
| 20 | 221.16446 | 8.28 | C13H20N2O | [M+H]+ | 2.83 | 0.00166 | 27.14 |
| 21 * | 221.10532 | 7.05 | C12H15NO3 | [M-H]− | 2.23 | 0.02531 | 3.44 |
| 22 * | 227.05528 | 7.65 | C8H9N3O5 | [M-H]− | 2.79 | 0.03370 | 0.03 |
| 23 | 229.19067 | 9.96 | C12H24N2O2 | [M+H]+ | 2.25 | 7.581 × 10−6 | 8.29 |
| 24 | 229.19089 | 10.34 | C12H24N2O2 | [M+H]+ | 2.31 | 0.00180 | 9.12 |
| 25 | 235.12117 | 7.97 | C11H16N4O2 | [M-H]− | 2.80 | 0.00063 | 14.47 |
| 26 | 267.20618 | 6.22 | C15H26N2O2 | [M+H]+ | 2.09 | 0.02326 | 14.27 |
| 27 * | 287.09198 | 10.33 | C15H9N7 | [M-H]− | 1.89 | 0.00209 | 6.52 |
| 28 | 290.12280 | 6.09 | C12H19NO7 | [M+H]+ | 1.99 | 0.02156 | 3.09 |
| 29 * | 313.14261 | 13.90 | C17H19N3O3 | [M+H]+ | 2.24 | 1.702 × 10−6 | 0.13 |
| 30 | 349.23645 | 11.38 | C21H32O4 | [M+H]+ | 1.93 | 0.01758 | 6.26 |
| 31 * | 363.21713 | 11.45 | C21H32O5 | [M-H]− | 2.30 | 0.00212 | 6.25 |
| 32 * | 365.23264 | 11.37 | C21H34O5 | [M-H]− | 2.32 | 0.00449 | 9.11 |
| 33 | 373.28003 | 7.04 | C18H36N4O4 | [M+H]+ | 1.90 | 0.01461 | 8.88 |
| 34 | 385.22260 | 11.11 | C20H34O7 | [M-H]− | 2.15 | 0.00248 | 6.78 |
| 35 * | 401.31131 | 7.963 | C20H40N4O4 | [M+H]+ | 2.34 | 0.00425 | 24.45 |
| 36 * | 423.29312 | 7.96 | C17H38N6O6 | [M+H]+ | 2.08 | 0.01770 | 44.49 |
| 37 | 609.32690 | 11.13 | C33H44N4O7 | [M+H]+ | 1.88 | 0.02610 | 2.75 |
| Rank | |||
|---|---|---|---|
| In-House Identifier (ID) | First | Second | Third |
| 1 | Amylamine | 2-Aminopentane | Isoamylamine |
| 2 | N-Cyclobutylacetamide | N-(3-Butenyl)acetamide | N-Ethyloxolan-2-imine |
| 3 | No candidate structures could be retrieved for the assigned chemical formula | ||
| 4 | Non-live (PubChem CID 57449799) | Non-live (PubChem CID 57424791) | Non-live (PubChem CID 57424804) |
| 5 | 4-Acetamido-1-pentanol | N-[(S)-1-(Hydroxymethyl)butyl]acetamide | N-[(2R,4R)-4-Hydroxypentan-2-yl]acetamide |
| 6 | No candidate structures could be retrieved for the assigned chemical formula | ||
| 7 | 2-Carbamoyl-2-cyanocyclopropane-1-carboxylic acid | 4-Nitrophenylhydroxylamine | 4-Amino-2-nitrophenol |
| 8 | No candidate structures could be retrieved for the assigned chemical formula | ||
| 9 | N-(Prop-2-enoylamino)oxyprop-2-enamide | 4(3H)-Pyrimidinone, 6-hydroxy-2-methoxy-5-methyl- | [(Z)-C-Ethenyl-N-hydroxycarbonimidoyl] (1E)-N-hydroxyprop-2-enimidate |
| 10 | Oxoverdazyl | - | - |
| 11 | N-(1-Hydroxybutan-2-yl)-2-methylpropanamide | Ethyl [isopropyl(methyl)amino]acetat | 2-(Hydroxymethyl)-N-propan-2-ylbutanamide |
| 12 | 1-(3-Hydroxyphenyl)propane-1,2-dione | 3-(3,5-Dihydroxyphenyl)prop-2-enal | (E)-3-(2,5-Dihydroxyphenyl)prop-2-enal |
| 13 | No candidate structures could be retrieved for the assigned chemical formula | ||
| 14 | N,N-Dimethylimidazo[1,2-b]pyrazole-5-carboxamide | N,N-Dimethylimidazo[1,2-b]pyrazole-1-carboxamide | N-[(Dimethylamino)methylene]pyrazine-2-carboxamide |
| 15 | No candidate structures could be retrieved for the assigned chemical formula | ||
| 16 | No candidate structures could be retrieved for the assigned chemical formula | ||
| 17 | N-[2-(1H-Imidazol-5-yl)ethyl]-2,2-dimethylpropanamide | N-[2-(1H-Imidazol-5-yl)ethyl]pentanamide | Dolichotheline |
| 18 | (2S)-2-Amino-N-butan-2-yl-4-methylsulfanylbutanamide | (2S)-2-Amino-N-(4-methylsulfanylbutan-2-yl)butanamide | (2S)-2-Amino-N-(2-methylpropyl)-4-methylsulfanylbutanamide |
| 19 | N-(6-Acetamidohexyl)propanamide | N-(7-Acetamidoheptyl)acetamide | N-(4-Acetamidobutyl)pentanamide |
| 20 | 2-Amino-N-(1-phenylpropan-2-yl)butanamide | N-Sec-Butyl-L-phenylalaninamide | (2S)-2-Amino-N-(1-phenylbutan-2-yl)propanamide |
| 21 | No relevant fragmentation data could be acquired, for which structural elucidation was not possible | ||
| 22 | No relevant fragmentation data could be acquired, for which structural elucidation was not possible | ||
| 23 | N-[(1R,4R)-4-(Propionylamino)-1-methylpentyl]propionamide | N-[5-(Propanoylamino)hexyl]propanamide | Leucyl-l-leucinal |
| 24 | 6-Acetamido-N-(2-methylpropyl)hexanamide | 6-Acetamido-N-butan-2-ylhexanamide | N-(5-Acetamidooctyl)acetamide |
| 25 | Non-live (PubChem CID 83431936) | Non-live (PubChem CID 83421718) | N-(2-Acetamidoethyl)-2-(methylamino)pyridine-3-carboxamide |
| 26 | 5-Butyl-1,5-diisocyanatononane | 1,9-Diisocyanato-5-methyl-5-propylnonane | 2-[2-[Di(propan-2-yl)amino]ethoxy]-6-methoxyaniline |
| 27 | No relevant fragmentation data could be acquired, for which structural elucidation was not possible | ||
| 28 | N-[(5R,6R,7S,8R)-6,7,8,9-Tetrahydroxy-2-methyl-3,4-dioxonon-1-en-5-yl]acetamide | [(2R,3S,4R,5R)-5-Acetamido-4-acetyloxy-2-hydroxy-6-oxohexan-3-yl] acetate | Triacetylmycosamine |
| 29 | No relevant fragmentation data could be acquired, for which structural elucidation was not possible | ||
| 30 | 7-[(1R,2S,5R)-2-Hydroxy-5-[(3S)-3-hydroxy-4-methyloct-1-en-6-ynyl]cyclopentyl]hept-5-enoic acid | (Z)-7-[(1R,2S,5R)-2-Hydroxy-5-[(E)-3-hydroxy-3-methyloct-1-en-6-ynyl]cyclopentyl]hept-5-enoic acid | 6a-Carbaprostaglandin I3 |
| 31 | No relevant fragmentation data could be acquired, for which structural elucidation was not possible | ||
| 32 | No relevant fragmentation data could be acquired, for which structural elucidation was not possible | ||
| 33 | 6-[Bis[2-(2-methylpropylamino)-2-oxoethyl]amino]-N-hydroxyhexanamide | Leucylleucyllysine | 2-[[2-(2,6-Diaminohexanoylamino)-4-methylpentanoyl]amino]-4-methylpentanoic acid |
| 34 | (E)-4,5-Dihydroxy-11-[3-(methoxymethyl)-4-oxooxetan-2-yl]-2,3,5,7-tetramethylundec-2-enoic acid | Methyl 11-(3-methoxymethyl-4-oxo-2-oxetanyl)-4,5-dihydroxy-3,5,7-trimethyl-2-undecenoate | 5,6-Dihydroxyprostaglandin E1 |
| 35 | No relevant fragmentation data could be acquired, for which structural elucidation was not possible | ||
| 36 | No relevant fragmentation data could be acquired, for which structural elucidation was not possible | ||
| 37 | No candidate structures could be retrieved for the assigned chemical formula | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamel Oroumieh, S.; Naserian, A.A.; Van Meulebroek, L.; De Paepe, E.; Valizadeh, R.; Vanhaecke, L. Metabolic Fingerprinting of Feces from Calves, Subjected to Gram-Negative Bacterial Endotoxin. Metabolites 2021, 11, 108. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11020108
Kamel Oroumieh S, Naserian AA, Van Meulebroek L, De Paepe E, Valizadeh R, Vanhaecke L. Metabolic Fingerprinting of Feces from Calves, Subjected to Gram-Negative Bacterial Endotoxin. Metabolites. 2021; 11(2):108. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11020108
Chicago/Turabian StyleKamel Oroumieh, Saeid, Abbas Ali Naserian, Lieven Van Meulebroek, Ellen De Paepe, Reza Valizadeh, and Lynn Vanhaecke. 2021. "Metabolic Fingerprinting of Feces from Calves, Subjected to Gram-Negative Bacterial Endotoxin" Metabolites 11, no. 2: 108. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo11020108