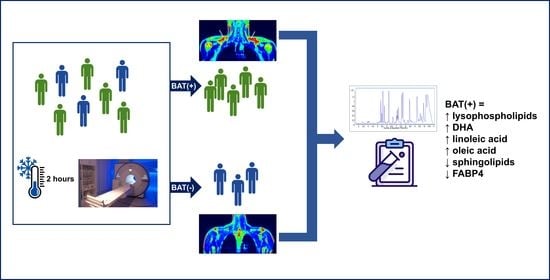
PET/MRI-Evaluated Activation of Brown Adipose Tissue via Cold Exposure Impacts Lipid Metabolism
Abstract
:1. Introduction
2. Results
2.1. Study Group Characteristics
2.2. Metabolic Fingerprinting of Plasma Samples
2.3. Pathway Analysis
| Metabolite | BAT Volume | BAT Activity | ||
|---|---|---|---|---|
| Correlation Coefficient | p-Value | Correlation Coefficient | p-Value | |
| L2 annotations | ||||
| A, Phospholipids | ||||
| LPC(14:0) | −0.45 | 0.04 | −0.48 | 0.02 |
| LPC(15:0) | −0.50 | 0.02 | −0.42 | 0.05 |
| LPC(16:0) | −0.34 | 0.1 | −0.46 | 0.03 |
| LPC(16:1) sn-2 | −0.55 | 0.009 | −0.41 | 0.06 |
| LPC(16:1) sn-1 | −0.42 | 0.05 | −0.61 | 0.003 |
| LPC(17:0) | −0.60 | 0.003 | −0.48 | 0.02 |
| LPC(17:1) | −0.43 | 0.04 | −0.31 | 0.2 |
| LPC(18:0) sn-2 | −0.58 | 0.005 | −0.50 | 0.02 |
| LPC(18:0) sn-1 | −0.59 | 0.004 | −0.57 | 0.006 |
| LPC(18:1) | −0.50 | 0.02 | −0.36 | 0.1 |
| LPC(18:2) | −0.65 | 0.0009 | −0.46 | 0.03 |
| LPC(19:0) | −0.63 | 0.002 | −0.58 | 0.005 |
| LPC(19:1) | −0.47 | 0.03 | −0.30 | 0.2 |
| LPC(20:0) | −0.30 | 0.2 | −0.51 | 0.01 |
| LPC(20:2) | −0.62 | 0.002 | −0.51 | 0.02 |
| LPC(20:4) | −0.62 | 0.002 | −0.43 | 0.05 |
| LPC(O-18:1)/LPC(P-18:0) sn-2 | −0.58 | 0.005 | −0.47 | 0.03 |
| LPC(O-18:1)/LPC(P-18:0) sn-1 | −0.47 | 0.03 | −0.35 | 0.1 |
| LPC(O-18:2)/LPC(P-18:1) | −0.67 | 0.0006 | −0.43 | 0.05 |
| LPC(16:0)-OH | −0.53 | 0.01 | −0.44 | 0.04 |
| LPE(18:0) | −0.50 | 0.02 | −0.50 | 0.02 |
| LPE(O-20:1)/LPE(P-20:0) | −0.49 | 0.02 | −0.45 | 0.03 |
| LPI(18:0) | −0.43 | 0.05 | −0.24 | 0.3 |
| PC(16:0_18:3) | −0.54 | 0.009 | −0.63 | 0.002 |
| PC(16:0_22:6) | −0.46 | 0.03 | −0.34 | 0.1 |
| PC(36:4) | −0.39 | 0.07 | −0.54 | 0.009 |
| PC(36:5) | −0.48 | 0.02 | −0.62 | 0.002 |
| PC(38:5) | −0.50 | 0.02 | −0.44 | 0.04 |
| PC(O-16:1_18:2)/ PC(P-16:0_18:2) | −0.43 | 0.04 | −0.35 | 0.1 |
| SM(d32:2) | −0.61 | 0.003 | −0.65 | 0.001 |
| SM(d35:2) | −0.53 | 0.01 | −0.40 | 0.06 |
| B, Other metabolites | ||||
| Histidine | −0.45 | 0.04 | −0.54 | 0.009 |
| L2/L3 annotations | ||||
| Bilirubin | −0.44 | 0.04 | −0.47 | 0.03 |
| DHA | 0.52 | 0.01 | 0.48 | 0.02 |
| Hexacosanedioic acid | −0.39 | 0.07 | −0.50 | 0.02 |
| FA 12:4;O3 | −0.59 | 0.003 | −0.51 | 0.02 |
| ST 19:1;O2;S (dihydrotestosterone sulfate) | −0.67 | 0.0007 | −0.51 | 0.02 |
2.4. Additional Analyses
3. Discussion
4. Materials and Methods
4.1. Study Group
4.2. Screening Visit
4.3. CE and PET-MR Scanning
4.4. Plasma Metabolic Fingerprinting
4.5. Serum Protein Measurement
4.6. Statistical Analysis
4.7. Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Europe Obesity. Available online: https://ec.europa.eu/eurostat/web/products-eurostat-news/-/ddn-20210721-2. (accessed on 30 March 2022).
- DeFronzo, R.A.; Bonadonna, R.C.; Ferrannini, E. Pathogenesis of NIDDM. A balanced overview. Diabetes Care 1992, 15, 318–368. [Google Scholar] [CrossRef]
- Ochner, C.N.; Barrios, D.M.; Lee, C.D.; Pi-Sunyer, F.X. Biological mechanisms that promote weight regain following weight loss in obese humans. Physiol. Behav. 2013, 120, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Maliszewska, K.; Kretowski, A. Brown Adipose Tissue and Its Role in Insulin and Glucose Homeostasis. Int. J. Mol. Sci. 2021, 22, 1530. [Google Scholar] [CrossRef]
- Fenzl, A.; Kiefer, F.W. Brown adipose tissue and thermogenesis. Horm. Mol. Biol. Clin. Investig. 2014, 19, 25–37. [Google Scholar] [CrossRef]
- Saely, C.H.; Geiger, K.; Drexel, H. Brown versus white adipose tissue: A mini-review. Gerontology 2012, 58, 15–23. [Google Scholar] [CrossRef]
- Cohade, C.; Osman, M.; Pannu, H.K.; Wahl, R.L. Uptake in supraclavicular area fat (“USA-Fat”): Description on 18F-FDG PET/CT. J. Nucl. Med. 2003, 44, 170–176. [Google Scholar]
- Yeung, H.W.; Grewal, R.K.; Gonen, M.; Schöder, H.; Larson, S.M. Patterns of (18)F-FDG uptake in adipose tissue and muscle: A potential source of false-positives for PET. J. Nucl. Med. 2003, 44, 1789–1796. [Google Scholar]
- Chen, L.; Lu, W.; Wang, L.; Xing, X.; Chen, Z.; Teng, X.; Zeng, X.; Muscarella, A.D.; Shen, Y.; Cowan, A.; et al. Metabolite discovery through global annotation of untargeted metabolomics data. Nat. Methods 2021, 18, 1377–1385. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Boon, M.R.; Bakker, L.E.H.; Prehn, C.; Adamski, J.; Vosselman, M.J.; Jazet, I.M.; Arias-Bouda, L.M.P.; van Lichtenbelt, W.D.M.; van Dijk, K.W.; Rensen, P.C.N.; et al. LysoPC-acyl C16:0 is associated with brown adipose tissue activity in men. Metabolomics 2017, 13, 48. [Google Scholar] [CrossRef] [Green Version]
- Xiang, A.S.; Giles, C.; Loh, R.K.C.; Formosa, M.F.; Eikelis, N.; Lambert, G.W.; Meikle, P.J.; Kingwell, B.A.; Carey, A.L. Plasma Docosahexaenoic Acid and Eicosapentaenoic Acid Concentrations Are Positively Associated with Brown Adipose Tissue Activity in Humans. Metabolites 2020, 10, 388. [Google Scholar] [CrossRef]
- Kovaničová, Z.; Karhánek, M.; Kurdiová, T.; Baláž, M.; Wolfrum, C.; Ukropcová, B.; Ukropec, J. Metabolomic Analysis Reveals Changes in Plasma Metabolites in Response to Acute Cold Stress and Their Relationships to Metabolic Health in Cold-Acclimatized Humans. Metabolites 2021, 11, 619. [Google Scholar] [CrossRef]
- Bargut, T.C.L.; Martins, F.F.; Santos, L.P.; Aguila, M.B.; Mandarim-de-Lacerda, C.A. Administration of eicosapentaenoic and docosahexaenoic acids may improve the remodeling and browning in subcutaneous white adipose tissue and thermogenic markers in brown adipose tissue in mice. Mol. Cell. Endocrinol. 2019, 482, 18–27. [Google Scholar] [CrossRef]
- Félix-Soriano, E.; Sáinz, N.; Gil-Iturbe, E.; Collantes, M.; Fernández-Galilea, M.; Castilla-Madrigal, R.; Ly, L.; Dalli, J.; Moreno-Aliaga, M.J. Changes in brown adipose tissue lipid mediator signatures with aging, obesity, and DHA supplementation in female mice. FASEB J. 2021, 35, e21592. [Google Scholar] [CrossRef]
- Lynes, M.D.; Leiria, L.O.; Lundh, M.; Bartelt, A.; Shamsi, F.; Huang, T.L.; Takahashi, H.; Hirshman, M.F.; Schlein, C.; Lee, A.; et al. The cold-induced lipokine 12,13-diHOME promotes fatty acid transport into brown adipose tissue. Nat. Med. 2017, 23, 631–637. [Google Scholar] [CrossRef] [Green Version]
- Błachnio-Zabielska, A.U.; Pułka, M.; Baranowski, M.; Nikołajuk, A.; Zabielski, P.; Górska, M.; Górski, J. Ceramide metabolism is affected by obesity and diabetes in human adipose tissue. J. Cell. Physiol. 2012, 227, 550–557. [Google Scholar] [CrossRef]
- Blachnio-Zabielska, A.U.; Koutsari, C.; Tchkonia, T.; Jensen, M.D. Sphingolipid content of human adipose tissue: Relationship to adiponectin and insulin resistance. Obesity 2012, 20, 2341–2347. [Google Scholar] [CrossRef] [Green Version]
- Ravichandran, S.; Finlin, B.S.; Kern, P.A.; Özcan, S. Sphk2. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 570–576. [Google Scholar] [CrossRef]
- Christoffersen, C.; Federspiel, C.K.; Borup, A.; Christensen, P.M.; Madsen, A.N.; Heine, M.; Nielsen, C.H.; Kjaer, A.; Holst, B.; Heeren, J.; et al. The Apolipoprotein M/S1P Axis Controls Triglyceride Metabolism and Brown Fat Activity. Cell Rep. 2018, 22, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Rajakumari, S.; Srivastava, S. Aging and β3-adrenergic stimulation alter mitochondrial lipidome of adipose tissue. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158922. [Google Scholar] [CrossRef]
- Wang, X.; Wahl, R. Responses of the insulin signaling pathways in the brown adipose tissue of rats following cold exposure. PLoS ONE 2014, 9, e99772. [Google Scholar] [CrossRef] [Green Version]
- Hanssen, M.J.; Hoeks, J.; Brans, B.; van der Lans, A.A.; Schaart, G.; van den Driessche, J.J.; Jörgensen, J.A.; Boekschoten, M.V.; Hesselink, M.K.; Havekes, B.; et al. Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus. Nat. Med. 2015, 21, 863–865. [Google Scholar] [CrossRef]
- Wang, T.Y.; Liu, C.; Wang, A.; Sun, Q. Intermittent cold exposure improves glucose homeostasis associated with brown and white adipose tissues in mice. Life Sci. 2015, 139, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Lin, Y.; Zhang, X.Y.; Wang, D.H. Cold exposure inhibits hypothalamic Kiss-1 gene expression, serum leptin concentration, and delays reproductive development in male Brandt’s vole (Lasiopodomys brandtii). Int. J. Biometeorol. 2015, 59, 679–691. [Google Scholar] [CrossRef]
- Ricci, M.R.; Fried, S.K.; Mittleman, K.D. Acute cold exposure decreases plasma leptin in women. Metabolism 2000, 49, 421–423. [Google Scholar] [CrossRef]
- Peinó, R.; Piñeiro, V.; Gualillo, O.; Menéndez, C.; Brenlla, J.; Casabiell, X.; Diéguez, C.; Casanueva, F.F. Cold exposure inhibits leptin secretion in vitro by a direct and non-specific action on adipose tissue. Eur. J. Endocrinol. 2000, 142, 195–199. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.; Yamamoto, A.; Watanabe, M.; Kataoka, M.; Terada, H.; Shinohara, Y. Quantitative evaluation of the effects of cold exposure of rats on the expression levels of ten FABP isoforms in brown adipose tissue. Biotechnol. Lett. 2011, 33, 237–242. [Google Scholar] [CrossRef]
- Vergnes, L.; Chin, R.; Young, S.G.; Reue, K. Heart-type fatty acid-binding protein is essential for efficient brown adipose tissue fatty acid oxidation and cold tolerance. J. Biol. Chem. 2011, 286, 380–390. [Google Scholar] [CrossRef] [Green Version]
- Shu, L.; Hoo, R.L.; Wu, X.; Pan, Y.; Lee, I.P.; Cheong, L.Y.; Bornstein, S.R.; Rong, X.; Guo, J.; Xu, A. A-FABP mediates adaptive thermogenesis by promoting intracellular activation of thyroid hormones in brown adipocytes. Nat. Commun. 2017, 8, 14147. [Google Scholar] [CrossRef] [Green Version]
- Stachoń, A.J. Menstrual Changes in Body Composition of Female Athletes. Coll. Antropol. 2016, 40, 111–122. [Google Scholar]
- Maliszewska, K.; Adamska-Patruno, E.; Miniewska, K.; Bauer, W.; Mojsak, M.; Kretowski, A. PET/MRI-evaluated brown adipose tissue activity may be related to dietary MUFA and omega-6 fatty acids intake. Sci. Rep. 2022, 12, 4112. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, K.A.; Peltoniemi, P.; Marjamäki, P.; Asola, M.; Strindberg, L.; Parkkola, R.; Huupponen, R.; Knuuti, J.; Lönnroth, P.; Nuutila, P. Human adipose tissue glucose uptake determined using [(18)F]-fluoro-deoxy-glucose ([(18)F]FDG) and PET in combination with microdialysis. Diabetologia 2001, 44, 2171–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniluk, U.; Daniluk, J.; Kucharski, R.; Kowalczyk, T.; Pietrowska, K.; Samczuk, P.; Filimoniuk, A.; Kretowski, A.; Lebensztejn, D.; Ciborowski, M. Untargeted Metabolomics and Inflammatory Markers Profiling in Children with Crohn’s Disease and Ulcerative Colitis-A Preliminary Study. Inflamm. Bowel Dis. 2019, 25, 1120–1128. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]






| Parameter | BAT(+) Median (Q1–Q3) | BAT(−) Median (Q1–Q3) | p-Value | |
|---|---|---|---|---|
| N | 17 | 8 | ||
| BAT activity [µmol × (100 g−1) × min−1] | 29.9 (23.6–37.8) | NA | ||
| BAT volume [mm3] | 15,280 (1859–31,490) | NA | ||
| Age [years] | 24 (23–26.5) | 27 (25–32.3) | 0.07 | |
| BMI [kg/m2] | 24.5 (22.6–28.7) | 26.7 (23.2–31.1) | 0.37 | |
| % AT [%] | 16.5 (13.3–22.0) | 22.0 (16.0–25.9) | 0.34 | |
| % SMM [%] | 47.5 (44.2–49.7) | 44.5 (42.3–47.0) | 0.22 | |
| VAT volume [cm3] | 420 (251–761.5) | 775.5 (297.8–1275) | 0.29 | |
| VAT mass [g] | 396 (237.5–718.5) | 731.5 (291.8–1203.3) | 0.24 | |
| HbA1c [mmol/L] | 4.9 (4.8–5.2) | 5.1 (4.9–5.1) | 0.42 | |
| OGTT | glc 0′ [μIU/L] | 97 (90.8–98.8) | 94 (91.5–95.5) | 0.25 |
| AUC glc | 183 (171.3–199.5) | 175 (166.5–193.5) | 0.45 | |
| ins 0′ [μIU/L] | 7.6 (6.3–11.8) | 8.8 (7.4–10.9) | 0.48 | |
| AUC ins | 22.4 (14.7–33.4) | 26.1 (21.9–51.9) | 0.19 | |
| Cholesterol [mg/dL] | 155 (135–177.5) | 174 (148.3–182) | 0.40 | |
| LDL-cholesterol [mg/dL] | 77.6 (65.9–111.5) | 101.5 (77.3–106.5) | 0.41 | |
| HDL-cholesterol [mg/dL] | 52 (39–73) | 57.5 (47.5–72.5) | 0.62 | |
| Triglycerides [mg/dL] | 86 (46.5–110) | 83 (46.3–108.5) | 0.98 | |
| CRP [mg/L] | 0.7 (0.3–2.7) | 0.7 (0.4–1.1) | 0.93 | |
| Creatinine [mg/dL] | 1 (0.9–1.1) | 0.9 (0.9–1.0) | 0.56 | |
| ALT [IU/L] | 21.8 (16.3–26.6) | 17.8 (15.7–28.3) | 0.54 | |
| AST [IU/L] | 20.4 (18.8–25.4) | 23.8 (18.3–31.0) | 0.66 | |
| TSH [μIU/mL] | 1.9 (1.5–3.2) | 197 (1.4–2.5) | 0.51 | |
| BAT(−) | BAT(+) | ||||||
|---|---|---|---|---|---|---|---|
| 60′ vs. 0′ | 120′ vs. 0′ | 240′ vs. 0′ | 60′ vs. 0′ | 120′ vs. 0′ | 240′ vs. 0′ | ||
| Glucose | Change [%] | 3.5 | −2.4 | −1.1 | 3.4 | 2.4 | 1.9 |
| p-value | 0.2 | 0.4 | 0.7 | 0.005 | 0.04 | 0.1 | |
| Insulin | Change [%] | −23.5 | −23.4 | −35.2 | −11.4 | −6.6 | −16.9 |
| p-value | 0.02 | 0.04 | 0.008 | 0.09 | 0.008 | 0.04 | |
| TNF-α | Change [%] | 41.9 | 59.4 | 50.6 | 48.8 | 12.3 | 45.9 |
| p-value | 0.2 | 0.04 | 0.2 | 0.2 | 0.3 | 0.1 | |
| FGF21 | Change [%] | −27.1 | −24.1 | −39.0 | −25.4 | −10.3 | −35.1 |
| p-value | 0.5 | 0.5 | 0.02 | 0.7 | 0.3 | 0.08 | |
| Leptin | Change [%] | −12.8 | −13.1 | −20.7 | −15.7 | −25.4 | −35.4 |
| p-value | 0.02 | 0.02 | 0.02 | 0.0007 | 3 × 10−5 | 0.0001 | |
| FABP4 | Change [%] | 5.1 | −9.8 | −22.6 | −12.9 | −30.7 | −20.1 |
| p-value | 1 | 0.5 | 0.5 | 0.003 | 5 × 10−5 | 0.0002 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miniewska, K.; Maliszewska, K.; Pietrowska, K.; Godzień, J.; Łabieniec, Ł.; Mojsak, M.; Krętowski, A.; Ciborowski, M. PET/MRI-Evaluated Activation of Brown Adipose Tissue via Cold Exposure Impacts Lipid Metabolism. Metabolites 2022, 12, 456. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12050456
Miniewska K, Maliszewska K, Pietrowska K, Godzień J, Łabieniec Ł, Mojsak M, Krętowski A, Ciborowski M. PET/MRI-Evaluated Activation of Brown Adipose Tissue via Cold Exposure Impacts Lipid Metabolism. Metabolites. 2022; 12(5):456. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12050456
Chicago/Turabian StyleMiniewska, Katarzyna, Katarzyna Maliszewska, Karolina Pietrowska, Joanna Godzień, Łukasz Łabieniec, Małgorzata Mojsak, Adam Krętowski, and Michał Ciborowski. 2022. "PET/MRI-Evaluated Activation of Brown Adipose Tissue via Cold Exposure Impacts Lipid Metabolism" Metabolites 12, no. 5: 456. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12050456