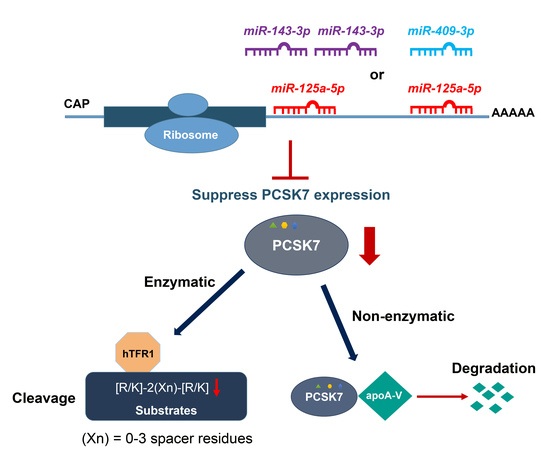
Post-Transcriptional Effects of miRNAs on PCSK7 Expression and Function: miR-125a-5p, miR-143-3p, and miR-409-3p as Negative Regulators
Abstract
:1. Introduction
2. Results
2.1. Bioinformatics Prediction of PCSK7-Targeting miRNAs
2.2. Expression of the Predicted miRNAs and PCSK7 in HEK293T, HepG2, and Huh7 Cells
2.3. Negative Regulation of PCSK7 mRNA Expression Levels by the Overexpression of miR-125a-5p, miR-143-3p, and miR-409-3p
2.4. Direct Interaction between the Predicted miRNAs and the 3′-UTR of PCSK7
2.5. Direct Interactions between miR-125a-5p, miR-143-3p, and miR-409-3p with the 3′-UTR of PCSK7
2.6. Functional Effects of miR-125a-5p on PC7 Activity
3. Discussion
4. Materials and Methods
4.1. Computational Prediction of miRNAs That Target the PCSK7 Gene
4.2. Cell Culture
4.3. RNA Extraction, Complementary DNA (cDNA) Synthesis, and Quantitative Polymerase Chain Reaction (qPCR)
4.4. Plasmids and Cell Transfection
4.4.1. miR-Overexpressing Vectors
4.4.2. Vectors Containing the PCSK7 3′-UTR Wild-Type and Mutated Forms
4.5. Luciferase Reporter Assay
4.6. Western Blot Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seidah, N.G.; Prat, A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 2012, 11, 367–383. [Google Scholar] [CrossRef] [PubMed]
- Seidah, N.G.; Hamelin, J.; Mamarbachi, M.; Dong, W.; Tardos, H.; Mbikay, M.; Chretien, M.; Day, R. cDNA structure, tissue distribution, and chromosomal localization of rat PC7, a novel mammalian proprotein convertase closest to yeast kexin- like proteinases. Proc. Natl. Acad. Sci. USA 1996, 93, 3388–3393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munzer, J.S.; Basak, A.; Zhong, M.; Mamarbachi, A.; Hamelin, J.; Savaria, D.; Lazure, C.; Hendy, G.N.; Benjannet, S.; Chretien, M.; et al. In vitro characterization of the novel proprotein convertase PC7. J. Biol. Chem. 1997, 272, 19672–19681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidah, N.G. Proprotein convertases PC4, PACE4, and PC7. In Handbook of the Biologically Active Peptides, 2nd ed.; Kastin, A.J., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 1812–1820. [Google Scholar]
- Van de Loo, J.W.; Creemers, J.W.; Bright, N.A.; Young, B.D.; Roebroek, A.J.; Van de Ven, W.J. Biosynthesis, distinct post-translational modifications, and functional characterization of lymphoma proprotein convertase. J. Biol. Chem. 1997, 272, 27116–27123. [Google Scholar] [CrossRef] [Green Version]
- Van de Loo, J.W.; Teuchert, M.; Pauli, I.; Plets, E.; Van de Ven, W.J.; Creemers, J.W. Dynamic palmitoylation of lymphoma proprotein convertase prolongs its half-life, but is not essential for trans-Golgi network localization. Biochem. J. 2000, 352, 827–833. [Google Scholar] [CrossRef]
- Declercq, J.; Meulemans, S.; Plets, E.; Creemers, J.W. Internalization of proprotein convertase PC7 from plasma membrane is mediated by a novel motif. J. Biol. Chem. 2012, 287, 9052–9060. [Google Scholar] [CrossRef] [Green Version]
- Guillemot, J.; Canuel, M.; Essalmani, R.; Prat, A.; Seidah, N.G. Implication of the proprotein convertases in iron homeostasis: Proprotein convertase 7 sheds human transferrin receptor 1 and furin activates hepcidin. Hepatology 2013, 57, 2514–2524. [Google Scholar] [CrossRef]
- Durand, L.; Duval, S.; Evagelidis, A.; Guillemot, J.; Dianati, V.; Sikorska, E.; Schu, P.; Day, R.; Seidah, N.G. The motif EXEXXXL in the cytosolic tail of the secretory human proprotein convertase PC7 regulates its trafficking and cleavage activity. J. Biol. Chem. 2020, 295, 2068–2083. [Google Scholar] [CrossRef]
- Ginefra, P.; Filippi, B.G.H.; Donovan, P.; Bessonnard, S.; Constam, D.B. Compartment-Specific Biosensors Reveal a Complementary Subcellular Distribution of Bioactive Furin and PC7. Cell Rep. 2018, 22, 2176–2189. [Google Scholar] [CrossRef] [Green Version]
- Rousselet, E.; Benjannet, S.; Hamelin, J.; Canuel, M.; Seidah, N.G. The Proprotein Convertase PC7: Unique zymogen activation and trafficking pathways. J. Biol. Chem. 2011, 286, 2728–2738. [Google Scholar] [CrossRef] [Green Version]
- Wetsel, W.C.; Rodriguiz, R.M.; Guillemot, J.; Rousselet, E.; Essalmani, R.; Kim, I.H.; Bryant, J.C.; Marcinkiewicz, J.; Desjardins, R.; Day, R.; et al. Disruption of the expression of the proprotein convertase PC7 reduces BDNF production and affects learning and memory in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 17362–17367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, Y.; Duval, S.; Sachan, V.; Essalmani, R.; Susan-Resiga, D.; Roubtsova, A.; Hamelin, J.; Gerhardy, S.; Kirchhofer, D.; Tagliabracci, V.S.; et al. Proprotein convertase 7 (PCSK7) reduces apoA-V levels. FEBS J. 2020, 287, 3565–3578. [Google Scholar] [CrossRef] [PubMed]
- Duval, S.; Abu-Thuraia, A.; Elkholi, I.E.; Chen, R.; Seebun, D.; Mayne, J.; Côté, J.F.; Figeys, D.; Seidah, N.G. Shedding of cancer susceptibility candidate 4 by the convertases PC7/furin unravels a novel secretory protein implicated in cancer progression. Cell Death Dis. 2020, 11, 665. [Google Scholar] [CrossRef] [PubMed]
- Rousselet, E.; Benjannet, S.; Marcinkiewicz, E.; Asselin, M.C.; Lazure, C.; Seidah, N.G. Proprotein convertase PC7 enhances the activation of the EGF receptor pathway through processing of the EGF precursor. J. Biol. Chem. 2011, 286, 9185–9195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dongiovanni, P.; Paolini, E.; Corsini, A.; Sirtori, C.R.; Ruscica, M. NAFLD or MAFLD diagnoses and cardiovascular diseases: From epidemiology to drug approaches. Eur. J. Clin. Investig. 2021, 51, e13519. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Meroni, M.; Baselli, G.; Mancina, R.M.; Ruscica, M.; Longo, M.; Rametta, R.; Cespiati, A.; Pelusi, S.; Ferri, N.; et al. PCSK7 gene variation bridges atherogenic dyslipidemia with hepatic inflammation in NAFLD patients. J. Lipid Res. 2019, 60, 1144–1153. [Google Scholar] [CrossRef]
- Buch, S.; Sharma, A.; Ryan, E.; Datz, C.; Griffiths, W.J.H.; Way, M.; Buckley, T.W.M.; Ryan, J.D.; Stewart, S.; Wright, C.; et al. Variants in PCSK7, PNPLA3 and TM6SF2 are risk factors for the development of cirrhosis in hereditary haemochromatosis. Aliment. Pharmacol. Ther. 2021, 53, 830–843. [Google Scholar] [CrossRef]
- Kawashiri, M.A. Can PCSK7 be A New Pharmaceutical Target? J. Atheroscler. Thromb. 2022, 29, ED188. [Google Scholar] [CrossRef]
- Naeli, P.; Mirzadeh Azad, F.; Malakootian, M.; Seidah, N.G.; Mowla, S.J. Post-transcriptional Regulation of PCSK9 by miR-191, miR-222, and miR-224. Front. Genet. 2017, 8, 189. [Google Scholar] [CrossRef] [Green Version]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Azad, F.M.; Arabian, M.; Maleki, M.; Malakootian, M. Small molecules with big impacts on cardiovascular diseases. Biochem. Genet. 2020, 58, 359–383. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, J.; Gao, T.; Xu, Z.; Cui, J. Dynamic and modularized microRNA regulation and its implication in human cancers. Sci. Rep. 2017, 7, 13356. [Google Scholar] [CrossRef] [Green Version]
- Van Solingen, C.; Oldebeken, S.R.; Salerno, A.G.; Wanschel, A.; Moore, K.J. High-Throughput Screening Identifies MicroRNAs Regulating Human PCSK9 and Hepatic Low-Density Lipoprotein Receptor Expression. Front. Cardiovasc. Med. 2021, 8, 667298. [Google Scholar] [CrossRef]
- Kang, S.; Zhao, Y.; Hu, K.; Xu, C.; Wang, L.; Liu, J.; Yao, A.; Zhang, H.; Cao, F. miR-124 exhibits antiproliferative and antiaggressive effects on prostate cancer cells through PACE4 pathway. Prostate 2014, 74, 1095–1106. [Google Scholar] [CrossRef]
- Loveday, E.-K.; Diederich, S.; Pasick, J.; Jean, F. Human microRNA-24 modulates highly pathogenic avian-origin H5N1 influenza A virus infection in A549 cells by targeting secretory pathway furin. J. Gen. Virol. 2015, 96, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Seidah, N.G.; Sadr, M.S.; Chretien, M.; Mbikay, M. The multifaceted Proprotein Convertases: Their unique, redundant, complementary and opposite functions. J. Biol. Chem. 2013, 288, 21473–21481. [Google Scholar] [CrossRef] [Green Version]
- Peloso, G.M.; Auer, P.L.; Bis, J.C.; Voorman, A.; Morrison, A.C.; Stitziel, N.O.; Brody, J.A.; Khetarpal, S.A.; Crosby, J.R.; Fornage, M. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am. J. Hum. Genet. 2014, 94, 223–232. [Google Scholar] [CrossRef] [Green Version]
- Kurano, M.; Tsukamoto, K.; Kamitsuji, S.; Kamatani, N.; Hara, M.; Ishikawa, T.; Kim, B.-J.; Moon, S.; Kim, Y.J.; Teramoto, T. Genome-wide association study of serum lipids confirms previously reported associations as well as new associations of common SNPs within PCSK7 gene with triglyceride. J. Hum. Genet. 2016, 61, 427–433. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: MicroRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taheri Bajgan, E.; Gholipour, A.; Faghihi, M.; Mowla, S.J.; Malakootian, M. Linc-ROR has a Potential ceRNA Activity for OCT4A by Sequestering miR-335-5p in the HEK293T Cell Line. Biochem. Genet. 2022, 60, 1007–1024. [Google Scholar] [CrossRef]
- Pelucchi, S.; Galimberti, S.; Greni, F.; Rametta, R.; Mariani, R.; Pelloni, I.; Girelli, D.; Busti, F.; Ravasi, G.; Valsecchi, M.G.; et al. Proprotein convertase 7 rs236918 associated with liver fibrosis in Italian patients with HFE-related hemochromatosis. J. Gastroenterol. Hepatol. 2016, 31, 1342–1348. [Google Scholar] [CrossRef] [PubMed]
- Josson, S.; Gururajan, M.; Sung, S.; Hu, P.; Shao, C.; Zhau, H.; Liu, C.; Lichterman, J.; Duan, P.; Li, Q. Stromal fibroblast-derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene 2015, 34, 2690–2699. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Tamizkar, K.H.; Hussen, B.M.; Taheri, M. MicroRNA signature in liver cancer. Pathol. -Res. Pract. 2021, 219, 153369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Lin, Y.; Sun, F.; Gao, J.; Han, B.; Li, S. miR-409 down-regulates Jak-Stat pathway to inhibit progression of liver cancer. Eur. Rev. Med. Pharmacol. Sci 2019, 23, 146–154. [Google Scholar] [CrossRef]
- Mamdouh, S.; Khorshed, F.; Aboushousha, T.; Hamdy, H.; Diab, A.; Seleem, M.; Saber, M. Evaluation of Mir-224, Mir-215 and Mir-143 as Serum Biomarkers for HCV Associated Hepatocellular Carcinoma. Asian Pac. J. Cancer Prev. 2017, 18, 3167–3171. [Google Scholar] [CrossRef]
- Zheng, J.; Zhou, Z.; Xu, Z.; Li, G.; Dong, P.; Chen, Z.; Lin, D.; Chen, B.; Yu, F. Serum microRNA-125a-5p, a useful biomarker in liver diseases, correlates with disease progression. Mol. Med. Rep. 2015, 12, 1584–1590. [Google Scholar] [CrossRef] [Green Version]
- Coppola, N.; Potenza, N.; Pisaturo, M.; Mosca, N.; Tonziello, G.; Signoriello, G.; Messina, V.; Sagnelli, C.; Russo, A.; Sagnelli, E. Liver microRNA hsa-miR-125a-5p in HBV chronic infection: Correlation with HBV replication and disease progression. PLoS ONE 2013, 8, e65336. [Google Scholar] [CrossRef]
- Tessitore, A.; Cicciarelli, G.; Del Vecchio, F.; Gaggiano, A.; Verzella, D.; Fischietti, M.; Mastroiaco, V.; Vetuschi, A.; Sferra, R.; Barnabei, R. MicroRNA expression analysis in high fat diet-induced NAFLD-NASH-HCC progression: Study on C57BL/6J mice. BMC Cancer 2016, 16, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Wu, H.; Zhang, H.; Fang, S.; Zeng, R. miR-143-3p inhibits proliferation and invasion of hepatocellular carcinoma cells by regulating its target gene FGF1. Clin. Transl. Oncol. 2021, 23, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-q.; Meng, H.; Wang, N.; Liang, L.-n.; Liu, L.-n.; Lu, S.-m.; Luan, Y. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn. Pathol. 2014, 9, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, H.; Chen, D.; Cai, C.; Du, Q.; Lin, H.; Pan, T.; Sheng, L.; Xu, Y.; Teng, T.; Tu, J. microRNA-143-3p attenuated development of hepatic fibrosis in autoimmune hepatitis through regulation of TAK1 phosphorylation. J. Cell. Mol. Med. 2020, 24, 1256–1267. [Google Scholar] [CrossRef] [Green Version]
- Tryndyak, V.P.; Marrone, A.K.; Latendresse, J.R.; Muskhelishvili, L.; Beland, F.A.; Pogribny, I.P. MicroRNA changes, activation of progenitor cells and severity of liver injury in mice induced by choline and folate deficiency. J. Nutr. Biochem. 2016, 28, 83–90. [Google Scholar] [CrossRef]
- Wu, H.; Song, X.; Ling, Y.; Zhou, J.; Tao, Z.; Shen, Y. Comprehensive bioinformatics analysis of critical lncRNAs, mRNAs and miRNAs in non-alcoholic fatty liver disease. Mol. Med. Rep. 2019, 19, 2649–2659. [Google Scholar] [CrossRef] [Green Version]
- Becker-Greene, D.; Li, H.; Perez-Cremades, D.; Wu, W.; Bestepe, F.; Ozdemir, D.; Niosi, C.E.; Aydogan, C.; Orgill, D.P.; Feinberg, M.W. MiR-409-3p targets a MAP4K3-ZEB1-PLGF signaling axis and controls brown adipose tissue angiogenesis and insulin resistance. Cell. Mol. Life Sci. 2021, 78, 7663–7679. [Google Scholar] [CrossRef]
- Oexle, K.; Ried, J.S.; Hicks, A.A.; Tanaka, T.; Hayward, C.; Bruegel, M.; Gögele, M.; Lichtner, P.; Müller-Myhsok, B.; Döring, A. Novel association to the proprotein convertase PCSK7 gene locus revealed by analysing soluble transferrin receptor (sTfR) levels. Hum. Mol. Genet. 2011, 20, 1042–1047. [Google Scholar] [CrossRef]
- Kramer, M.F. Stem-loop RT-qPCR for miRNAs. Curr. Protoc. Mol. Biol. 2011, 95, 15.10.1–15.10.15. [Google Scholar] [CrossRef]
- Guillemot, J.; Seidah, N.G. The proprotein convertase PC7 sheds the human TfR1 independent of HFE. Hepatology 2013, 58, 1861–1862. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malakootian, M.; Naeli, P.; Mowla, S.J.; Seidah, N.G. Post-Transcriptional Effects of miRNAs on PCSK7 Expression and Function: miR-125a-5p, miR-143-3p, and miR-409-3p as Negative Regulators. Metabolites 2022, 12, 588. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12070588
Malakootian M, Naeli P, Mowla SJ, Seidah NG. Post-Transcriptional Effects of miRNAs on PCSK7 Expression and Function: miR-125a-5p, miR-143-3p, and miR-409-3p as Negative Regulators. Metabolites. 2022; 12(7):588. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12070588
Chicago/Turabian StyleMalakootian, Mahshid, Parisa Naeli, Seyed Javad Mowla, and Nabil G. Seidah. 2022. "Post-Transcriptional Effects of miRNAs on PCSK7 Expression and Function: miR-125a-5p, miR-143-3p, and miR-409-3p as Negative Regulators" Metabolites 12, no. 7: 588. https://0-doi-org.brum.beds.ac.uk/10.3390/metabo12070588