Automated Protein Secondary Structure Assignment from Cα Positions Using Neural Networks
Abstract
:1. Introduction
- (i)
- Methods that assign SSE directly from local geometric parameters derived from C positions, e.g., local distances along the chain (DEFINE_S, SABA, STICK), possibly combined with information about close spatial neighbors (P-SEA), dihedral angles (PALSSE), and contact map (VoTAP); fitting a curve to C points was also utilized [6,7].
- (ii)
- Methods that cut the query structure into short C structural fragments and compare them to structural fragments extracted from known protein structures, e.g., by means of trained Bayesian (SST) or a Nearest-Neighbor Classifier (SACF).
- (iii)
- Methods that use Machine Learning (ML) to infer SSE. The progress in the field of ML observed in the twenty-first century significantly increased the popularity of these methods in bioinformatics in general, including the problem of the secondary structure assignment. The PCASSO [14] method uses the Random Forest classifier technique, where 16 features are solely based on C coordinates. It reports a very high accuracy reaching 96% with respect to DSSP. Another Random Forest-based approach RaFoSA [15] uses 30 features: 1 × residue type, 6 × C-C distances, the angle between three C atoms, 4 × torsional angles formed by four C atoms, and the number of C-C contacts. Similarly, it is also 96% accurate with respect to DSSP. More sophisticated methods employ Neural Networks [16] and Convolutional Neural Networks (CNN), as observed in the DLFSA [17]. The accuracy reported by the authors in the latter case is somewhat lower: around 83% depending on the PDB files. A consensus approach was also described, where the final assignment was decided as a consensus of four different ML techniques [18].
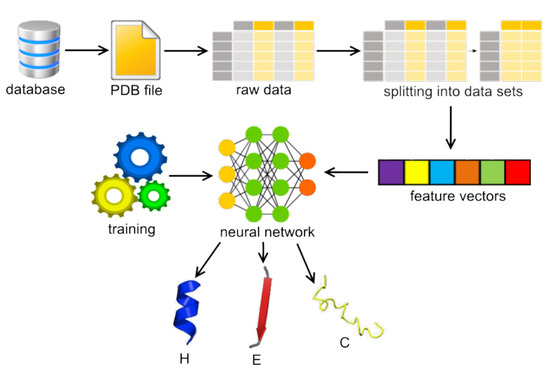
2. Methods
2.1. Network Architecture
2.2. Software Implementation
2.3. Data Sets
2.4. Input Features
- (i)
- Local , , and , distances measured between C atoms along a contiguous segment, referred to as local in the text (see Figure 2A). The is a signed value, i.e., it is the distance between atoms; i and are multiplied by the sign of . This allows the classifier to distinguish between left-handed and right-handed conformations. All possible such distances were calculated within a segment, e.g., for five , four , and three were included in the input tensor.
- (ii)
- The number of spatial neighbors found around each C atom of a segment within a given distance, referred to as neighbors in the text (see Figure 2B). Four distance cutoffs were used: 4, 4.5, 5, and 6 Å. Therefore, for , the input tensor includes contact counts. For example, in Figure 2B, the middle C atom (darkest gray) has eight neighbors within a 5 Å radius (medium-dark). Atoms separated by at most two residues along the sequence are also not included in the count (light gray).
- (iii)
- The number of hydrogen bonds formed by each C atom of a segment, referred to as hbonds in the text. A coarse-grained hydrogen-bonding model, which has recently been developed in our laboratory (submitted) for the SURPASS algorithm [44,45], was used for detecting such bonds (see Figure 2C). Detailed derivation and assessment of the potential will be published elsewhere; a summary of the algorithm is provided in the Supporting Materials. In brief, a triangle is constructed from every three subsequent C atoms. According to our coarse-grained definition, a given triangle may form a hydrogen bond with another triangle when specific geometry criteria are met, i.e., when the two triangles are roughly parallel. Therefore, the criteria for such a hydrogen-bonding event are based on the mutual orientation of two local coordinate systems constructed on the two C triangles. Respective geometric criteria were derived from PDB statistics so that they match the all-atom hydrogen bonds observed in the PDB deposits. According to our model, each residue located in a beta-strand may form up to two such hydrogen bonds on either side of the triangle. Therefore, the HECA input tensor for a fragment of N residues contains N integer hydrogen bond counts that are either 0, 1, or 2.
3. Results and Discussion
4. Summary
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins: Two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Frishman, D.; Argos, P. Knowledge-based protein secondary structure assignment. Proteins 1995, 23, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Richards, F.M.; Kundrot, C.E. Identification of structural motifs from protein coordinate data: Secondary structure and first-level supersecondary structure. Proteins Struct. Funct. Bioinform. 1988, 3, 71–84. [Google Scholar] [CrossRef]
- Sklenar, H.; Etchebest, C.; Lavery, R. Describing protein structure: A general algorithm yielding complete helicoidal parameters and a unique overall axis. Proteins Struct. Funct. Bioinform. 1989, 6, 46–60. [Google Scholar] [CrossRef]
- Hosseini, S.R.; Sadeghi, M.; Pezeshk, H.; Eslahchi, C.; Habibi, M. PROSIGN: A method for protein secondary structure assignment based on three-dimensional coordinates of consecutive Cα atoms. Comput. Biol. Chem. 2008, 32, 406–411. [Google Scholar] [CrossRef]
- Cao, C.; Wang, G.; Liu, A.; Xu, S.; Wang, L.; Zou, S. A New Secondary Structure Assignment Algorithm Using Cα Backbone Fragments. Int. J. Mol. Sci. 2016, 17, 333. [Google Scholar] [CrossRef] [Green Version]
- Labesse, G.; Colloc’h, N.; Pothier, J.; Mornon, J.R. P-sea: A new efficient assignment of secondary structure from cαl trace of proteins. Bioinformatics 1997, 13, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Majumdar, I.; Krishna, S.S.; Grishin, N.V. PALSSE: A program to delineate linear secondary structural elements from protein structures. BMC Bioinform. 2005, 6, 202. [Google Scholar] [CrossRef] [Green Version]
- Taylor, W.R. Defining linear segments in protein structure. J. Mol. Biol. 2001, 310, 1135–1150. [Google Scholar] [CrossRef] [Green Version]
- Dupuis, F.; Sadoc, J.F.; Mornon, J.P. Protein Secondary Structure Assignment Through Voronoï Tessellation. Proteins Struct. Funct. Genet. 2004, 55, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Yoo, M.J.; Shin, J.; Cho, K.H. SABA (secondary structure assignment program based on only alpha carbons): A novel pseudo center geometrical criterion for accurate assignment of protein secondary structures. BMB Rep. 2011, 44, 118–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konagurthu, A.S.; Lesk, A.M.; Allison, L. Minimum message length inference of secondary structure from protein coordinate data. Bioinformatics 2012, 28, i97–i105. [Google Scholar] [CrossRef] [PubMed]
- Law, S.M.; Frank, A.T.; Brooks, C.L. PCASSO: A fast and efficient Cα-based method for accurately assigning protein secondary structure elements. J. Comput. Chem. 2014, 35, 1757–1761. [Google Scholar] [CrossRef] [Green Version]
- Salawu, E.O. RaFoSA: Random forests secondary structure assignment for coarse-grained and all-atom protein systems. Cogent Biol. 2016, 2, 1214061. [Google Scholar] [CrossRef]
- Nasr, K.A.; Sekmen, A.; Bilgin, B.; Jones, C.; Koku, A.B. Deep Learning for Assignment of Protein Secondary Structure Elements from C Coordinates. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 2546–2552. [Google Scholar] [CrossRef]
- Antony, J.V.; Madhu, P.; Balakrishnan, J.P.; Yadav, H. Assigning secondary structure in proteins using AI. J. Mol. Model. 2021, 27, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Sallal, M.A.; Chen, W.; Nasr, K.A. Machine Learning Approach to Assign Protein Secondary Structure Elements from Ca Trace. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea, 16–19 December 2020; pp. 35–41. [Google Scholar] [CrossRef]
- Levitt, M.; Warshel, A. Computer simulation of protein folding. Nature 1975, 253, 694–698. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Dustin Schaeffer, R.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Sieradzan, A.K.; Czaplewski, C.; Krupa, P.; Mozolewska, M.A.; Karczyńska, A.S.; Lipska, A.G.; Lubecka, E.A.; Gołaś, E.; Wirecki, T.; Makowski, M.; et al. Modeling the Structure, Dynamics, and Transformations of Proteins with the UNRES Force Field; Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2022; Volume 2376, pp. 399–416. [Google Scholar] [CrossRef]
- Vicatos, S.; Rychkova, A.; Mukherjee, S.; Warshel, A. An effective Coarse-grained model for biological simulations: Recent refinements and validations. Proteins Struct. Funct. Bioinform. 2014, 82, 1168–1185. [Google Scholar] [CrossRef]
- Monticelli, L.; Kandasamy, S.K.; Periole, X.; Larson, R.G.; Tieleman, D.P.; Marrink, S.J. The MARTINI coarse-grained force field: Extension to proteins. J. Chem. Theory Comput. 2008, 4, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Tieleman, D.P. Perspective on the martini model. Chem. Soc. Rev. 2013, 42, 6801–6822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liwo, A.; Czaplewski, C.; Sieradzan, A.K.; Lipska, A.G.; Samsonov, S.A.; Murarka, R.K. Theory and Practice of Coarse-Grained Molecular Dynamics of Biologically Important Systems. Biomolecules 2021, 11, 1347. [Google Scholar] [CrossRef]
- Wu, H.; Wolynes, P.G.; Papoian, G.A. AWSEM-IDP: A Coarse-Grained Force Field for Intrinsically Disordered Proteins. J. Phys. Chem. B 2018, 122, 11115–11125. [Google Scholar] [CrossRef] [PubMed]
- Tesei, G.; Schulze, T.K.; Crehuet, R.; Lindorff-Larsen, K. Accurate model of liquid-liquid phase behavior of intrinsically disordered proteins from optimization of single-chain properties. Proc. Natl. Acad. Sci. USA 2021, 118, e2111696118. [Google Scholar] [CrossRef]
- Kurcinski, M.; Badaczewska-Dawid, A.; Kolinski, M.; Kolinski, A.; Kmiecik, S. Flexible docking of peptides to proteins using CABS-dock. Protein Sci. 2020, 29, 211–222. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.; Jung, J.; Kobayashi, C.; Torre, D.U.L.; Takada, S.; Sugita, Y. Implementation of residue-level coarsegrained models in GENESIS for large-scale molecular dynamics simulations. PLoS Comput. Biol. 2022, 18, e1009578. [Google Scholar] [CrossRef]
- Kulik, M.; Mori, T.; Sugita, Y. Multi-Scale Flexible Fitting of Proteins to Cryo-EM Density Maps at Medium Resolution. Front. Mol. Biosci. 2021, 8, 61. [Google Scholar] [CrossRef]
- Kolinski, A.; Gront, D. Comparative modeling without implicit sequence alignments. Bioinformatics 2007, 23, 2522–2527. [Google Scholar] [CrossRef] [Green Version]
- Davtyan, A.; Schafer, N.P.; Zheng, W.; Clementi, C.; Wolynes, P.G.; Papoian, G.A. AWSEM-MD: Protein structure prediction using coarse-grained physical potentials and bioinformatically based local structure biasing. J. Phys. Chem. B 2012, 116, 8494–8503. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Ahlstrom, L.S.; Brooks, C.L. Exploring Protein–Nanoparticle Interactions with Coarse-Grained Protein Folding Models. Small 2017, 13, 1603748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzzo, A.; Delarue, P.; Rojas, A.; Nicolaï, A.; Maisuradze, G.G.; Senet, P. Missense Mutations Modify the Conformational Ensemble of the α-Synuclein Monomer Which Exhibits a Two-Phase Characteristic. Front. Mol. Biosci. 2021, 8, 6123. [Google Scholar] [CrossRef] [PubMed]
- Liwo, A.; Czaplewski, C.; Sieradzan, A.K.; Lubecka, E.A.; Lipska, A.G.; Golon, Ł.; Karczynska, A.; Krupa, P.; Mozolewska, M.A.; Makowski, M.; et al. Scale-consistent approach to the derivation of coarse-grained force fields for simulating structure, dynamics, and thermodynamics of biopolymers. Prog. Mol. Biol. Transl. Sci. 2020, 170, 73–122. [Google Scholar] [CrossRef] [PubMed]
- Kůrková, V. Kolmogorov’s theorem and multilayer neural networks. Neural Netw. 1992, 5, 501–506. [Google Scholar] [CrossRef]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Macnar, J.M.; Szulc, N.A.; Kryś, J.D.; Badaczewska-Dawid, A.E.; Gront, D. BioShell 3.0: Library for Processing Structural Biology Data. Biomolecules 2020, 10, 461. [Google Scholar] [CrossRef] [Green Version]
- Kryś, J.D.; Gront, D. VisuaLife: Library for interactive visualization in rich web applications. Bioinformatics 2021, 37, 3662–3663. [Google Scholar] [CrossRef]
- Wang, G.; Dunbrack, R.L. PISCES: A protein sequence culling server. Bioinformatics 2003, 19, 1589–1591. [Google Scholar] [CrossRef] [Green Version]
- Gront, D.; Kolinski, A. BioShell - a package of tools for structural biology computations. Bioinformatics 2006, 22, 621–622. [Google Scholar] [CrossRef] [Green Version]
- Gront, D.; Kolinski, A. Utility library for structural bioinformatics. Bioinformatics 2008, 24, 584–585. [Google Scholar] [CrossRef] [Green Version]
- Dawid, A.; Gront, D.; Kolinski, A. SURPASS Low-Resolution Coarse-Grained Protein Modeling. J. Chem. Theory Comput. 2017, 13, 5766–5779. [Google Scholar] [CrossRef] [PubMed]
- Dawid, A.; Gront, D.; Kolinski, A. Coarse-Grained Modeling of the Interplay between Secondary Structure Propensities and Protein Fold Assembly. J. Chem. Theory Comput. 2018, 14, 2277–2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kmiecik, S.; Gront, D.; Kolinski, M.; Wieteska, L.; Dawid, A.; Kolinski, A. Coarse-Grained Protein Models and Their Applications. Chem. Rev. 2016, 116, 7898–7936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wabik, J.; Kmiecik, S.; Gront, D.; Kouza, M.; Koliński, A. Combining coarse-grained protein models with replica-exchange all-atom molecular dynamics. Int. J. Mol. Sci. 2013, 14, 9893–9905. [Google Scholar] [CrossRef]
- Gront, D.; Kmiecik, S.; Kolinski, A. Backbone building from quadrilaterals: A fast and accurate algorithm for protein backbone reconstruction from alpha carbon coordinates. J. Comput. Chem. 2007, 28, 1593–1597. [Google Scholar] [CrossRef]




| Fragment Length | Local | Local + Neighbors | Local + Neighbors + Hbonds | |||
|---|---|---|---|---|---|---|
| Training | Validation | Training | Validation | Training | Validation | |
| 5 | 83.58 | 83.60 | 88.93 | 88.75 | 91.91 | 91.98 |
| 7 | 89.89 | 89.99 | 96.20 | 96.39 | 95.40 | 95.48 |
| 9 | 91.84 | 91.88 | 94.03 | 94.10 | 96.85 | 96.91 |
| 11 | 92.37 | 92.46 | 94.51 | 94.57 | 97.29 | 97.33 |
| 13 | 92.53 | 92.61 | 94.68 | 94.89 | 97.39 | 97.48 |
| Fragment length | 5 | 7 | 9 | 11 | 13 |
| No. of ideally predicted proteins | 144 | 246 | 406 | 465 | 491 |
| Differences between predicted and true classes | H: 6.97% | H: 2.87% | H: 2.78% | H: 2.05% | H: 1.88% |
| E: 5.16% | E: 4.55% | E: 4.30% | E: 3.44% | E: 3.63% | |
| C: 2.27% | C: 7.69% | C: 7.69% | C: 4.50% | C: 4.44% | |
| Average differences | 8.13% | 5.03% | 3.70% | 3.33% | 3.31% |
| 5-mer | Predicted | 11-mer | Predicted | ||||
|---|---|---|---|---|---|---|---|
| H | E | C | H | E | C | ||
| H | 93.026% | 0.035% | 6.938% | H | 97.953% | 0.036% | 2.009% |
| E | 0.180% | 94.840% | 4.978% | E | 0.123% | 96.559% | 3.317% |
| C | 11.089% | 1.179% | 87.731% | C | 2.771% | 1.730% | 95.498% |
| 13-mer | Predicted | 13-mer | Predicted | ||||
| H | E | C | H | E | C | ||
| H | 98.11% | 0.028% | 1.859% | H | 97.680% | 0.205% | 2.114% |
| E | 0.134% | 96.37% | 3.49% | E | 0.684% | 85.680% | 13.634% |
| C | 2.80% | 1.634% | 95.563% | C | 3.398% | 7.043% | 89.557% |
| PDB | Nres | Type | HECA | PCASSO | Nasr et al. | PDB | Nres | Type | HECA | PCASSO | Nasr et al. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1EAR_A | 135 | 0.957 | 0.936 | 0.956 | 4MNC_A | 299 | + | 0.950 | 0.849 | 0.930 | |
| 1GQI_A | 702 | 0.971 | 0.879 | 0.925 | 4MYD_A | 246 | + | 0.972 | 0.857 | 0.947 | |
| 1NUY_A | 322 | + | 0.960 | 0.859 | 0.891 | 4OH7_A | 296 | 0.953 | 0.427 | 0.909 | |
| 1OK0_A | 68 | 0.972 | 0.972 | 0.926 | 4P3H_A | 184 | + | 0.921 | 0.473 | 0.918 | |
| 1SDI_A | 207 | 0.962 | 0.901 | 0.981 | 4WKA_A | 363 | + | 0.962 | 0.853 | 0.909 | |
| 1UJ8_A | 66 | 0.917 | 0.794 | 0.939 | 4ZDS_A | 125 | 0.939 | 0.825 | 0.960 | ||
| 1Z6N_A | 160 | 0.945 | 0.867 | 0.950 | 5CKL_A | 175 | 0.933 | 0.883 | 0.937 | ||
| 2FGQ_X | 324 | 0.954 | 0.921 | 0.920 | 5CL8_A | 225 | 0.948 | 0.852 | 0.951 | ||
| 2FP1_A | 159 | 0.963 | 0.890 | 0.981 | 5CVW_A | 142 | 0.946 | 0.773 | 0.831 | ||
| 2FVY_A | 298 | + | 0.977 | 0.898 | 0.953 | 5GZK_A | 412 | 0.935 | 0.861 | 0.876 | |
| 2I5V_O | 239 | 0.979 | 0.951 | 0.954 | 5JUH_A | 130 | + | 0.948 | 0.627 | 0.862 | |
| 2JDA_A | 131 | 0.920 | 0.791 | 0.756 | 5LT5_A | 198 | + | 0.960 | 0.877 | 0.909 | |
| 2O1T_A | 428 | + | 0.907 | 0.847 | 0.930 | 5T9Y_A | 312 | + | 0.915 | 0.598 | 0.933 |
| 2OPC_A | 109 | 0.956 | 0.930 | 0.908 | 5TIF_A | 176 | 0.961 | 0.839 | 0.955 | ||
| 2QKV_A | 85 | + | 0.923 | 0.945 | 0.976 | 5UEB_A | 136 | + | 0.943 | 0.829 | 0.956 |
| 2RIN_A | 282 | + | 0.895 | 0.788 | 0.901 | 5W53_A | 297 | 0.963 | 0.947 | 0.970 | |
| 2RIQ_A | 129 | 0.888 | 0.903 | 0.938 | 5WEC_A | 104 | 0.946 | 0.866 | 0.981 | ||
| 2Z6R_A | 256 | + | 0.958 | 0.852 | 0.930 | 5YDE_A | 105 | + | 0.936 | 0.891 | 0.924 |
| 2ZDP_A | 104 | + | 0.972 | 0.798 | 0.904 | 5ZIM_A | 222 | 0.942 | 0.912 | 0.959 | |
| 3BQP_A | 74 | 0.937 | 0.900 | 1.000 | 6A2W_A | 159 | 0.957 | 0.831 | 0.975 | ||
| 3D2Y_A | 251 | + | 0.968 | 0.914 | 0.952 | 6E7E_A | 163 | 0.970 | 0.905 | 0.982 | |
| 3DXY_A | 200 | + | 0.932 | 0.869 | 0.970 | 6ER6_A | 82 | 0.931 | 0.886 | 0.976 | |
| 3KYJ_A | 123 | 0.961 | 0.658 | 0.992 | 6GEH_A | 250 | + | 0.968 | 0.910 | 0.956 | |
| 3LFK_A | 115 | + | 0.909 | 0.811 | 0.930 | 6I1A_A | 352 | 0.946 | 0.731 | 0.932 | |
| 3NJN_A | 108 | 0.903 | 0.517 | 0.861 | 6IY4_I | 86 | + | 0.881 | 0.838 | 0.907 | |
| 3OBQ_A | 135 | + | 0.950 | 0.907 | 0.956 | 6JH9_B | 22 | 0.655 | 0.689 | 0.773 | |
| 3Q40_A | 169 | 0.964 | 0.946 | 0.975 | 6JM5_A | 114 | + | 0.918 | 0.886 | 0.904 | |
| 3R87_A | 125 | + | 0.984 | 0.916 | 0.960 | 6JU1_A | 387 | + | 0.961 | 0.785 | 0.925 |
| 3RT2_A | 165 | + | 0.959 | 0.935 | 0.964 | 6JWF_A | 400 | 0.973 | 0.899 | 0.915 | |
| 3V4K_A | 180 | + | 0.967 | 0.672 | 0.967 | 6KTK_A | 362 | + | 0.956 | 0.495 | 0.945 |
| 3VK5_A | 247 | + | 0.956 | 0.889 | 0.976 | 6NEY_A | 119 | 0.936 | 0.880 | 0.933 | |
| 3VMK_A | 363 | + | 0.924 | 0.523 | 0.898 | 6NZS_A | 581 | 0.950 | 0.873 | 0.880 | |
| 3WDN_A | 119 | + | 0.952 | 0.872 | 0.933 | 6P80_A | 312 | 0.952 | 0.874 | 0.942 | |
| 4AYO_A | 428 | 0.965 | 0.875 | 0.914 | 6TM6_A | 90 | 0.855 | 0.876 | 0.878 | ||
| 4B20_A | 264 | + | 0.950 | 0.662 | 0.930 | 6TZX_A | 217 | 0.950 | 0.865 | 0.926 | |
| 4GMU_A | 604 | + | 0.960 | 0.863 | 0.904 | 6ULO_A | 310 | 0.940 | 0.340 | 0.923 | |
| 4JUI_A | 463 | + | 0.970 | 0.704 | 0.935 | 6YDR_A | 122 | 0.976 | 0.860 | 0.992 | |
| 4L9E_A | 108 | + | 0.947 | 0.904 | 0.917 | average | 0.942 | 0.820 | 0.931 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saqib, M.N.; Kryś, J.D.; Gront, D. Automated Protein Secondary Structure Assignment from Cα Positions Using Neural Networks. Biomolecules 2022, 12, 841. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12060841
Saqib MN, Kryś JD, Gront D. Automated Protein Secondary Structure Assignment from Cα Positions Using Neural Networks. Biomolecules. 2022; 12(6):841. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12060841
Chicago/Turabian StyleSaqib, Mohammad N., Justyna D. Kryś, and Dominik Gront. 2022. "Automated Protein Secondary Structure Assignment from Cα Positions Using Neural Networks" Biomolecules 12, no. 6: 841. https://0-doi-org.brum.beds.ac.uk/10.3390/biom12060841