Molecular Insights into Poly(ADP-ribose) Recognition and Processing
Abstract
:1. Metabolism and Cellular Function of PAR

2. Structural and Functional Diversity of PAR-Binding Modules
2.1. PAR Binding Motif (PBM)
2.2. Poly(ADP-Ribose)-Binding Zinc Finger (PBZ)
2.3. WWE Domain
2.4. Macrodomain
3. PARG—A New Member of the Macrodomain Family of Proteins
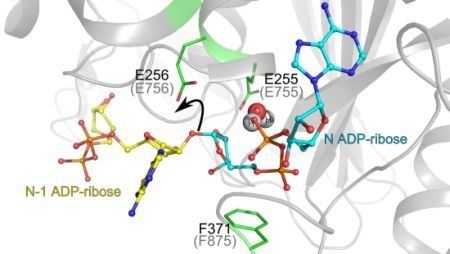
3.1. Catalytic Activity of PARG Enzymes


3.2. Regulation of PARG Enzymes
3.3. PARG as a Therapeutic Target
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Hassa, P.O.; Haenni, S.S.; Elser, M.; Hottiger, M.O. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Bio. Rev. 2006, 7, 789–829. [Google Scholar]
- Yates, S.P.; Jørgensen, R.; Andersen, G.R.; Merrill, R. Stealth and mimicry by deadly bacterial toxins. Trends Biochem. Sci. 2006, 31, 123–133. [Google Scholar] [CrossRef]
- Holbourn, K.P.; Shone, C.C.; Acharya, K.R. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006, 273, 4579–4593. [Google Scholar] [CrossRef]
- Di Girolamo, M.; Dani, N.; Stilla, A.; Corda, D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005, 272, 4565–4575. [Google Scholar] [CrossRef]
- Laing, S.; Unger, M.; Koch-Nolte, F.; Haag, F. ADP-ribosylation of arginine. Amino acids 2011, 41, 257–269. [Google Scholar] [CrossRef]
- Matic, I.; Ahel, I.; Hay, R.T. Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat. Methods 2012, 9, 771–772. [Google Scholar] [CrossRef]
- Amé, J.C.; Spenlehauer, C.; de Murcia, G. The PARP superfamily. BioEssays: news and reviews in molecular, cellular and developmental biology 2004, 26, 882–893. [Google Scholar] [CrossRef]
- Citarelli, M.; Teotia, S.; Lamb, R.S. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol. Biol. 2010, 10, 308. [Google Scholar] [CrossRef]
- Otto, H.; Reche, P. a; Bazan, F.; Dittmar, K.; Haag, F.; Koch-Nolte, F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 2005, 6, 139. [Google Scholar] [CrossRef]
- Slade, D.; Dunstan, M.S.; Barkauskaite, E.; Weston, R.; Lafite, P.; Dixon, N.; Ahel, M.; Leys, D.; Ahel, I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 2011, 477, 616–620. [Google Scholar] [CrossRef]
- Kraus, W.L. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008, 20, 294–302. [Google Scholar] [CrossRef]
- Sousa., F.G.; Matuo, R.; Soares, D.G.; Escargueil, A.E.; Henriques, J.A.P.; Larsen, A.K.; Saffi, J. PARPs and the DNA damage response. Carcinogenesis 2012, 33, 1433–1440. [Google Scholar] [CrossRef]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef]
- Kleine, H.; Poreba, E.; Lesniewicz, K.; Hassa, P.O.; Hottiger, M.O.; Litchfield, D.W.; Shilton, B.H.; Lüscher, B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 2008, 32, 57–69. [Google Scholar] [CrossRef]
- Aguiar, R.C.T.; Takeyama, K.; He, C.; Kreinbrink, K.; Shipp, M. A B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 2005, 280, 33756–33765. [Google Scholar]
- Riquelme, T.; Burzio, L.; Koide, S. ADP Ribosylation of Rat Liver Histone in vitro. J. Biol. Chem. 1979, 8, 3018–3028. [Google Scholar]
- Tao, Z.; Gao, P.; Liu, H.W. Identification of the ADP-Ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 2009, 131, 14258–14260. [Google Scholar] [CrossRef]
- Altmeyer, M.; Messner, S.; Hassa, P.O.; Fey, M.; Hottiger, M.O. Molecular mechanism of poly(ADP- ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009, 37, 3723–3738. [Google Scholar]
- Messner, S.; Altmeyer, M.; Zhao, H.; Pozivil, A.; Roschitzki, B.; Gehrig, P.; Rutishauser, D.; Huang, D.; Caflisch, A.; Hottiger, M.O. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010, 38, 6350–6362. [Google Scholar]
- Rolli, V.; O’Farrell, M.; Menissier-de Murcia, J.; de Murcia, G. Random mutagenesis of the poly(ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry 1997, 36, 12147–12154. [Google Scholar] [CrossRef]
- Juarez-Salinas, H.; Levi, V.; Jacobson, E.L.; Jacobson, M.K. Poly(ADP-ribose) has a branched structure in vivo. J. Biol. Chem. 1982, 257, 607–609. [Google Scholar]
- Kanai, M.; Miwa, M.; Kuchino, Y.; Sugimura, T. Presence of branched portion in poly(adenosine diphosphate ribose) in vivo. J. Biol. Chem. 1982, 257, 6217–6223. [Google Scholar]
- Miwa, M.; Saikawa, N.; Yamaizumi, Z.; Nishimura, S.; Sugimura, T. Structure of poly(adenosine diphosphate ribose): identification of 2'-[1''-ribosyl-2''-(or 3''-)(1'''-ribosyl)]adenosine-5',5'',5'''-tris(phosphate) as a branch linkage. Proc. Natl. Acad. Sci. USA 1979, 76, 595–599. [Google Scholar] [CrossRef]
- Ame, J.C.; Rolli, V.; Schreiber, V.; Niedergang, C.; Apiou, F.; Decker, P.; Muller, S.; Hoger, T.; Menissier-de Murcia, J.; de Murcia, G. PARP- 2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999, 274, 17860–17868. [Google Scholar]
- Rulten, S.L.; Fisher, A.E.O.; Robert, I.; Zuma, M.C.; Rouleau, M.; Ju, L.; Poirier, G.; Reina-San-Martin, B.; Caldecott, K.W. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell 2011, 41, 33–45. [Google Scholar] [CrossRef]
- Wielckens, K.; Schmidt, A.; George, E.; Bredehorst, R.; Hilz, H. DNA fragmentation and NAD depletion. Their relation to the turnover of endogenous mono(ADP-ribosyl) and poly(ADP-ribosyl) proteins. J. Biol. Chem. 1982, 257, 12872–12877. [Google Scholar]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef]
- Langelier, M.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage–dependent poly(ADP-ribosyl)ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef]
- Pleschke, J.M.; Kleczkowska, H.E.; Strohm, M.; Althaus, F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000, 275, 40974–40980. [Google Scholar]
- De Vos, M.; Schreiber, V.; Dantzer, F. The diverse roles and clinical relevance of PARPs in DNA damage repair: state of the art. Biochem. Pharmacol. 2012, 84, 137–146. [Google Scholar]
- Ahel, D.; Horejsí, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P.; Owen-Hughes, T.; Boulton, S.J. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar]
- Polo, S.E.; Kaidi, A.; Baskcomb, L.; Galanty, Y.; Jackson, S.P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010, 29, 3130–3139. [Google Scholar] [CrossRef]
- Mehrotra, P.V.; Ahel, D.; Ryan, D.P.; Weston, R.; Wiechens, N.; Kraehenbuehl, R.; Owen-Hughes, T.; Ahel, I. DNA repair factor APLF is a histone chaperone. Mol. Cell 2011, 41, 46–55. [Google Scholar] [CrossRef]
- Andrabi, S.A.; Kim, N.S.; Yu, S.W.; Wang, H.; Koh, D.W.; Sasaki, M.; Klaus, J.A.; Otsuka, T.; Zhang, Z.; Koehler, R.C. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA 2006, 103, 18308–18313. [Google Scholar]
- Chang, P.; Coughlin, M.; Mitchison, T.J. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005, 7, 1133–1139. [Google Scholar] [CrossRef]
- Chang, P.; Jacobson, M.K.; Mitchison, T.J. Poly(ADP-ribose) is required for spindle assembly and structure. Nature 2004, 432, 645–649. [Google Scholar] [CrossRef]
- Kim, M.K.; Dudognon, C.; Smith, S. Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep. 2012, 13, 724–732. [Google Scholar] [CrossRef]
- Hsiao, S.J.; Smith, S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 2008, 90, 83–92. [Google Scholar] [CrossRef]
- Muramatsu, Y.; Ohishi, T.; Sakamoto, M.; Tsuruo, T.; Seimiya, H. Cross-species difference in telomeric function of tankyrase 1. Cancer Sci. 2007, 98, 850–857. [Google Scholar] [CrossRef]
- Ahel, I.; Ahel, D.; Matsusaka, T.; Clark, A.J.; Pines, J.; Boulton, S.J.; West, S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 2008, 451, 81–85. [Google Scholar]
- Kashima, L.; Idogawa, M.; Mita, H.; Shitashige, M.; Yamada, T.; Ogi, K.; Suzuki, H.; Toyota, M.; Ariga, H.; Sasaki, Y.; Tokino, T. CHFR regulates the mitotic checkpoint by targeting PARP-1 for ubiquitination and degradation. J. Biol. Chem. 2012, 287, 12975–12984. [Google Scholar]
- Kothe, G.O.; Kitamura, M.; Masutani, M.; Selker, E.U.; Inoue, H. PARP is involved in replicative aging in Neurospora crassa. Fungal Genet. Biol. 2010, 47, 297–309. [Google Scholar] [CrossRef]
- Müller-Ohldach, M.; Brust, D.; Hamann, A.; Osiewacz, H.D. Overexpression of PaParp encoding the poly(ADP-ribose) polymerase of Podospora anserina affects organismal aging. Mech. Ageing Dev. 2011, 132, 33–42. [Google Scholar] [CrossRef]
- Alvarez-Gonzalez, R.; Althaus, F.R. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat. Res. 1989, 218, 67–74. [Google Scholar] [CrossRef]
- Lin, W.; Ame, J.C.; Aboul-Ela, N.; Jacobson, E.L.; Jacobson, M.K. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 1997, 272, 11895–11901. [Google Scholar] [CrossRef]
- Kim, I.K.; Kiefer, J.R.; Ho, C.M.W.; Stegeman, R. A; Classen, S.; Tainer, J. A; Ellenberger, T. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nat. Struct. Mol. Biol. 2012, 19, 653–656. [Google Scholar] [CrossRef]
- Dunstan, M.S.; Barkauskaite, E.; Lafite, P.; Knezevic, C.E.; Brassington, A.; Ahel, M.; Hergenrother, P.J.; Leys, D.; Ahel, I. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat. Commun. 2012, 3, 878. [Google Scholar] [CrossRef]
- Oka, S.; Kato, J.; Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006, 281, 705–713. [Google Scholar]
- Niere, M.; Mashimo, M.; Agledal, L.; Dölle, C.; Kasamatsu, A.; Kato, J.; Moss, J.; Ziegler, M. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose). J. Biol. Chem. 2012, 287, 16088–16102. [Google Scholar]
- Ono, T.; Kasamatsu, A.; Oka, S.; and Moss, J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl. Acad. Sci. USA 2006, 103, 16687–16691. [Google Scholar] [CrossRef]
- Tong, L.; Denu, J.M. Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. BBA-Proteins Proteom. 2010, 1804, 1617–1625. [Google Scholar] [CrossRef]
- Oka, J.; Ueda, K.; Hayaishi, O.; Komura, H.; Nakanishi, K. ADP-Ribosyl Protein Lyase. J. Biol. Chem. 1984, 259, 986–995. [Google Scholar]
- Gagné, J.P.; Isabelle, M.; Lo, K.S.; Bourassa, S.; Hendzel, M.J.; Dawson, V.L.; Dawson, T.M.; Poirier, G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008, 36, 6959–6976. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Kim, N.S.; Haince, J.; Kang, H.C.; David, K.K.; Andrabi, S.A.; Poirier, G.G.; Dawson, V.; Dawson, T.M. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci. Signal. 2011, 4, ra20. [Google Scholar] [CrossRef]
- Kleine, H.; Lüscher, B. Learning how to read ADP-ribosylation. Cell 2009, 139, 17–19. [Google Scholar] [CrossRef]
- Iles, N.; Rulten, S.; El-Khamisy, S.F.; Caldecott, K.W. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 2007, 27, 3793–3803. [Google Scholar] [CrossRef]
- Eustermann, S.; Brockmann, C.; Mehrotra, P.V.; Yang, J.C.; Loakes, D.; West, S.C.; Ahel, I.; Neuhaus, D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose). Nat. Struct. Mol. Biol. 2010, 17, 241–243. [Google Scholar] [CrossRef]
- Li, G.-Y.; McCulloch, R.D.; Fenton, A.L.; Cheung, M.; Meng, L.; Ikura, M.; Koch, C.A. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. USA 2010, 107, 9129–9134. [Google Scholar]
- Oberoi, J.; Richards, M.W.; Crumpler, S.; Brown, N.; Blagg, J.; Bayliss, R. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR). J. Biol. Chem. 2010, 285, 39348–39358. [Google Scholar]
- Aravind, L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 2001, 26, 273–275. [Google Scholar] [CrossRef]
- Callow, M.G.; Tran, H.; Phu, L.; Lau, T.; Lee, J.; Sandoval, W.N.; Liu, P.S.; Bheddah, S.; Tao, J.; Lill, J.R.; Hongo, J.A.; Davis, D.; Kirkpatrick, D.S.; Polakis, P.; Costa, M. Ubiquitin ligase RNF146 regulates tankyrase and axin to promote Wnt signaling. PLoS ONE 2011, 6, e22595. [Google Scholar]
- Kang, H.C.; Lee, Y.I.; Shin, J.H.; Andrabi, S.A.; Chi, Z.; Gagne, J.P.; Lee, Y.; Ko, H.S.; Lee, B.D.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA 2011, 108, 14103–14108. [Google Scholar]
- Zhang, Y.; Liu, S.; Mickanin, C.; Feng, Y.; Charlat, O.; Michaud, G.A.; Schirle, M.; Shi, X.; Hild, M.; Bauer, A.; Myer, V.E.; Finan, P.M.; Porter, J.A.; Huang, S.M.A.; Cong, F. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2009, 13, 623–629. [Google Scholar]
- Wang, Z.; Michaud, G.; Cheng, Z.; Zhang, Y.; Hinds, T.R.; Fan, E.; Cong, F.; Xu, W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012, 26, 235–240. [Google Scholar]
- Till, S.; Ladurner, A.G. Sensing NAD metabolites through macro domains. Front. Biosci-Landmark 2009, 14, 3246–3258. [Google Scholar]
- Karras, G.I.; Kustatscher, G.; Buhecha, H.R.; Allen, M.D.; Pugieux, C.; Sait, F.; Bycroft, M.; Ladurner, A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005, 24, 1911–1920. [Google Scholar] [CrossRef]
- Kustatscher, G.; Hothorn, M.; Pugieux, C.; Scheffzek, K; Ladurner, A.G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005, 12, 624–625. [Google Scholar] [CrossRef]
- Chen, D.; Vollmar, M.; Rossi, M.N.; Phillips, C.; Kraehenbuehl, R.; Slade, D.; Mehrotra, P.V.; von Delft, F.; Crosthwaite, S.K.; Gileadi, O.; Denu, J.M.; Ahel, I. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 2011, 286, 13261–13271. [Google Scholar]
- Peterson, F.C.; Chen, D.; Lytle, B.L.; Rossi, M.N.; Ahel, I.; Denu, J.M.; Volkman, B.F. Orphan macrodomain protein (Human C6orf130) is an O-Acyl-ADP-ribose deacylase: Solution structure and catalytic properties. J. Biol. Chem. 2011, 286, 35955–35965. [Google Scholar]
- Timinszky, G.; Till, S.; Hassa, P.O.; Hothorn, M.; Kustatscher, G.; Nijmeijer, B.; Colombelli, J.; Altmeyer, M.; Stelzer, E.H.; Scheffzek, K.; Hottiger, M.O.; Ladurner, A.G. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009, 16, 923–929. [Google Scholar] [CrossRef]
- Gottschalk, A.; Timinszky, G.; Kong, S.; Jin, J.; Cai, Y.; Swanson, S.; Washburn, M.; Florens, L.; Ladurner, A.; Conaway, J.W.; Conaway, R.C. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA 2009, 106, 13770–13774. [Google Scholar]
- Putics, Á.; Filipowicz, W.; Hall, J.; Alexander, E.; Ziebuhr, J.; Gorbalenya, A.E. ADP-ribose-1"-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005, 79, 12721–12731. [Google Scholar] [CrossRef]
- Egloff, M.P.; Malet, H.; Putics, A.; Heinonen, M.; Dutartre, H.; Frangeul, A.; Gruez, A.; Campanacci, V.; Cambillau, C.; Ziebuhr, J.; Ahola, T.; Canard, B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006, 80, 8493–8502. [Google Scholar]
- Shull, N.P.; Spinelli, S.L.; Phizicky, E.M. A highly specific phosphatase that acts on ADP-ribose 1’'-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 2005, 33, 650–660. [Google Scholar] [CrossRef]
- Botta, D.; Jacobson, M.K. Identification of a regulatory segment of poly(ADP-ribose) glycohydrolase. Biochemistry 2010, 49, 7674–7682. [Google Scholar] [CrossRef]
- Patel, C.N.; Koh, D.W.; Jacobson, M.K.; Oliveira, M.A. Identification of three critical acidic residues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARG catalytic domain. Biochem. J. 2005, 388, 493–500. [Google Scholar] [CrossRef]
- Panda, S.; Poirier, G.G.; Kay, S.A. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the Arabidopsis circadian oscillator. Dev. Cell 2002, 3, 51–61. [Google Scholar] [CrossRef]
- Brochu, G.; Duchaine, C.; Tribeault, L.; Lagueux, J.; Shah, G.M.; Poirier, G.G. Mode of action of poly(ADP-ribose) glycohydrolase. BBA-Gene Struct. Exp. 1994, 1219, 342–350. [Google Scholar]
- Mortusewicz, O.; Fouquerel, E.; Amé, J.C.; Leonhardt, H.; Schreiber, V. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 2011, 39, 5045–5056. [Google Scholar] [CrossRef]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef]
- Warbrick, E. PCNA binding through a conserved motif. Bioessays 1998, 20, 195–199. [Google Scholar] [CrossRef]
- Meyer, R.G.; Meyer-Ficca, M.L.; Whatcott, C.J.; Jacobson, E.L.; Jacobson, M.K. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp. Cell Res. 2007, 313, 2920–2936. [Google Scholar] [CrossRef]
- Meyer-Ficca, M.L.; Meyer, R.G.; Coyle, D.L.; Jacobson, E.L.; Jacobson, M.K. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 2004, 297, 521–532. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, H.; Lord, C.J.; Tutt, A.H.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; Martin, N.M.B.; Jackson, S.P.; Smith, G.C.M.; Ashworth, A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917–921. [Google Scholar]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O'Connor, M.J.; Ashworth, A.; Carmichael, J.; Kaye, S.B.; Schellens, J.H.M.; De Bono, J.S. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef]
- Hatakeyama, K.; Nemoto, Y.; Ueda, K.; Hayaishi, O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose). J. Biol. Chem. 1986, 261, 14902–14911. [Google Scholar]
- St-Laurent, J.F.; Gagnon, S.N.; Dequen, F.; Hardy, I.; Desnoyers, S. Altered DNA damage response in Caenorhabditis elegans with impaired poly(ADP-ribose) glycohydrolases genes expression. DNA Repair 2007, 6, 329–343. [Google Scholar]
- Ame, J.C.; Fouquerel, E.; Gauthier, L.R.; Biard, D.; Boussin, F.D.; Dantzer, F.; de Murcia, G.; Schreiber, V. Radiation-induced mitotic catastrophe in PARG-deficient cells. J. Cell Sci. 2009, 122, 1990–2002. [Google Scholar]
- Koh, D.W.; Lawler, A.M.; Poitras, M.F.; Sasaki, M.; Wattler, S.; Nehls, M.C.; Stoger, T.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA 2004, 101, 17699–17704. [Google Scholar]
- Zhou, Y.; Feng, X.; Koh, D.W. Enhanced DNA accessibility and increased DNA damage induced by the absence of poly(ADP-ribose) hydrolysis. Biochemistry 2010, 49, 7360–7366. [Google Scholar]
- Fathers, C.; Drayton, R.M.; Solovieva, S.; Bryant, H.E. Inhibition of poly(ADP-ribose) glycohydrolase (PARG) specifically kills BRCA2-deficient tumor cells. Cell Cycle 2012, 11, 990–997. [Google Scholar] [CrossRef]
- Slama, J.T.; Aboul-Ela, N.; Goli, D.M.; Cheesman, B.V.; Simmons, A.M.; Jacobson, M.K. Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J. Med. Chem. 1995, 38, 389–393. [Google Scholar]
- Finch, K.E.; Knezevic, C.E.; Nottbohm, A.C.; Partlow, K.C.; Hergenrother, P.J. Selective small molecule inhibition of poly(ADP-ribose) glycohydrolase (PARG). J. Am. Chem. Soc. 2012, 7, 563–570. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Žaja, R.; Mikoč, A.; Barkauskaite, E.; Ahel, I. Molecular Insights into Poly(ADP-ribose) Recognition and Processing. Biomolecules 2013, 3, 1-17. https://0-doi-org.brum.beds.ac.uk/10.3390/biom3010001
Žaja R, Mikoč A, Barkauskaite E, Ahel I. Molecular Insights into Poly(ADP-ribose) Recognition and Processing. Biomolecules. 2013; 3(1):1-17. https://0-doi-org.brum.beds.ac.uk/10.3390/biom3010001
Chicago/Turabian StyleŽaja, Roko, Andreja Mikoč, Eva Barkauskaite, and Ivan Ahel. 2013. "Molecular Insights into Poly(ADP-ribose) Recognition and Processing" Biomolecules 3, no. 1: 1-17. https://0-doi-org.brum.beds.ac.uk/10.3390/biom3010001