Cyclodextrins Can Entrap Zearalenone-14-Glucoside: Interaction of the Masked Mycotoxin with Cyclodextrins and Cyclodextrin Bead Polymer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Fluorescence Spectroscopic Measurements
2.3. Modeling Studies
2.3.1. Pharmacophoric Analysis of the CD Cavity
2.3.2. Docking Study
2.4. Extraction of Z14G from Aqueous Solution by BBP
2.5. HPLC Analysis
2.6. Statistical Analyses
3. Results
3.1. Fluorescence Excitation and Emission Spectra of Z14G
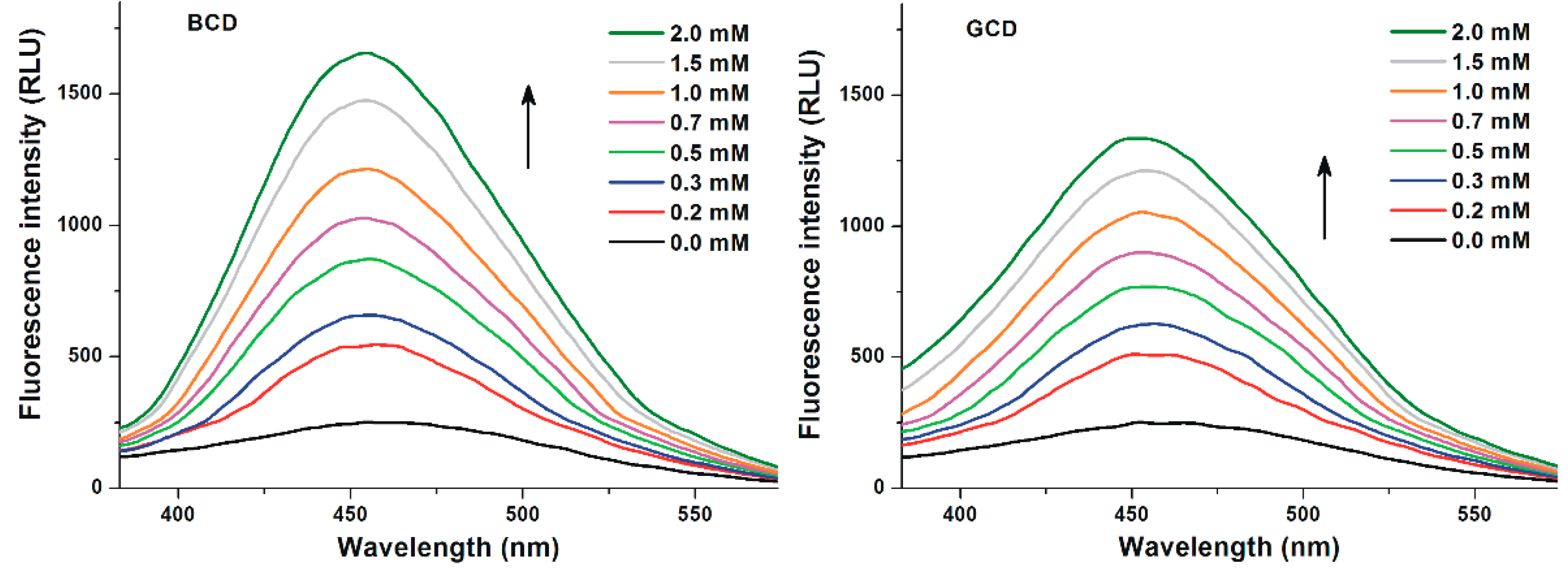
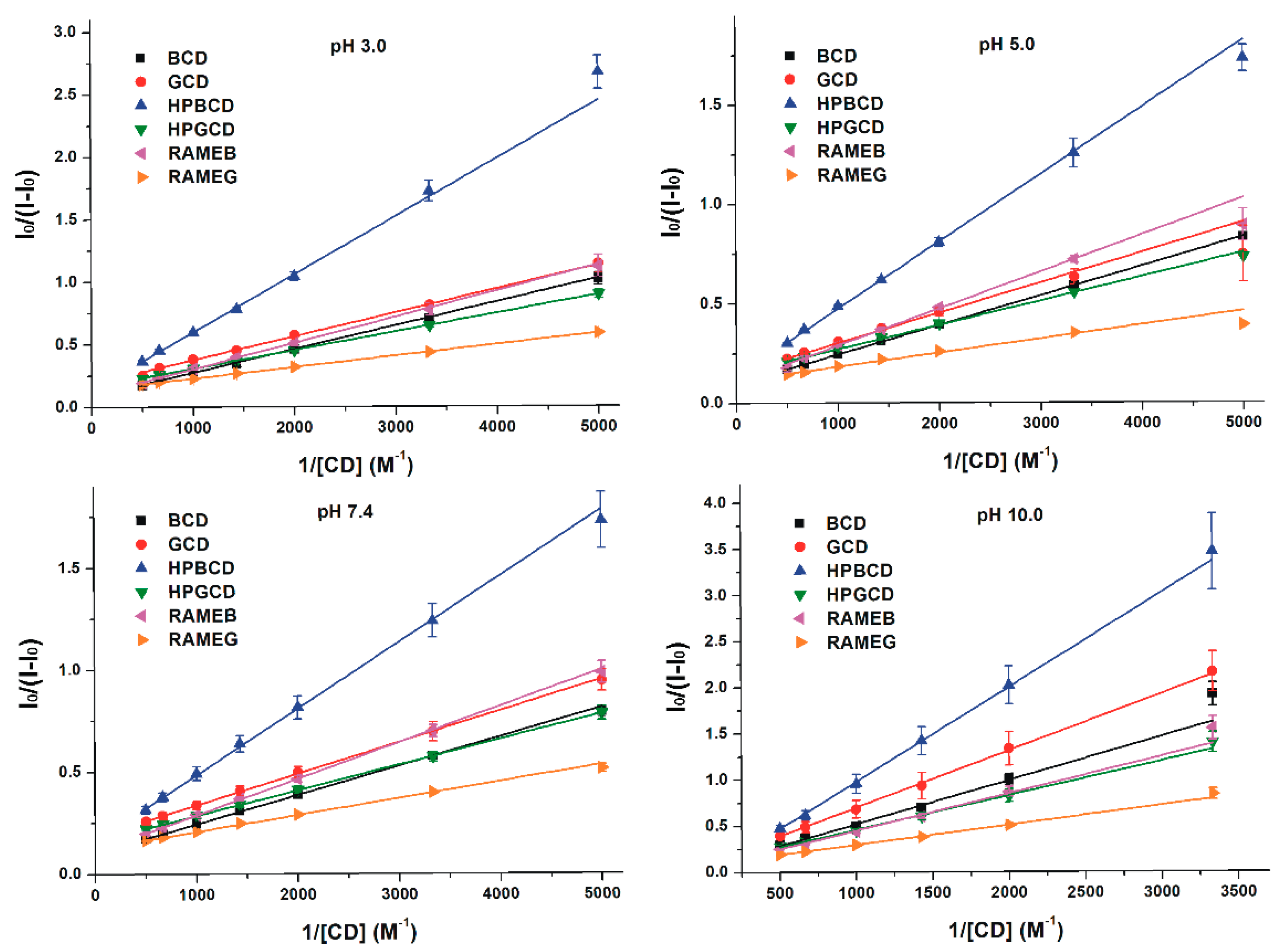
3.2. Effects of Cyclodextrins on the Fluorescence Signal of Z14G
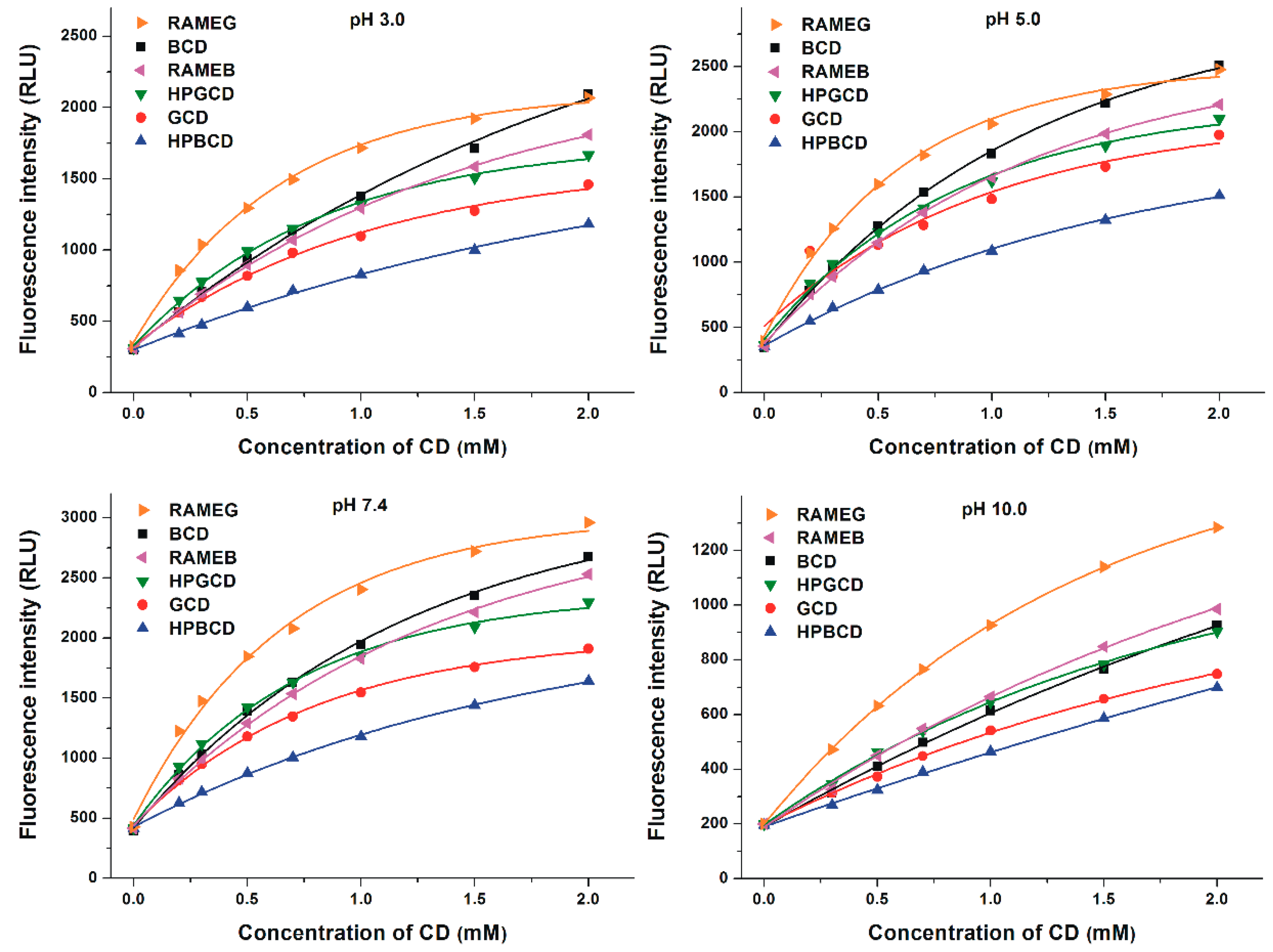
3.3. Binding Constants of Z14G-CD Complexes
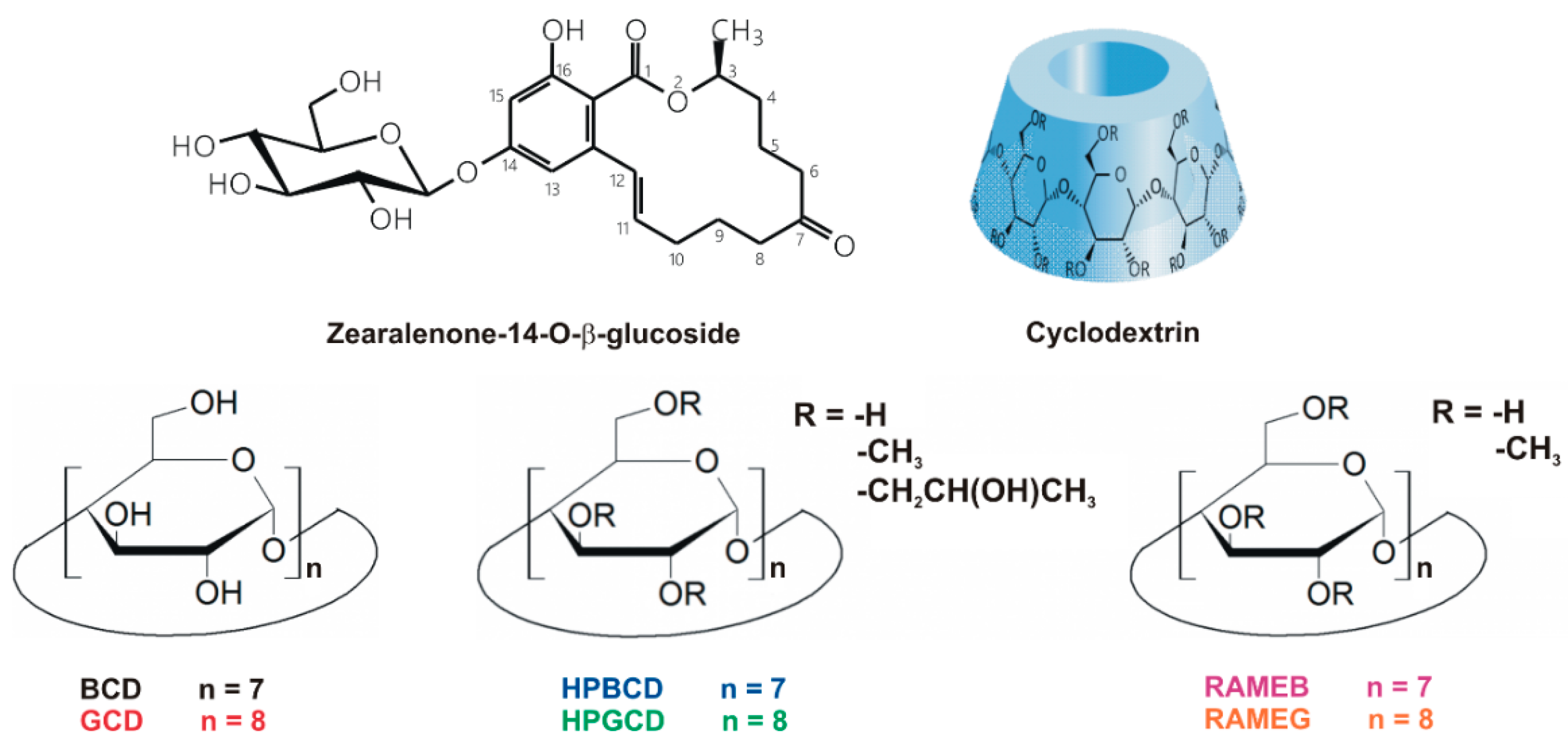
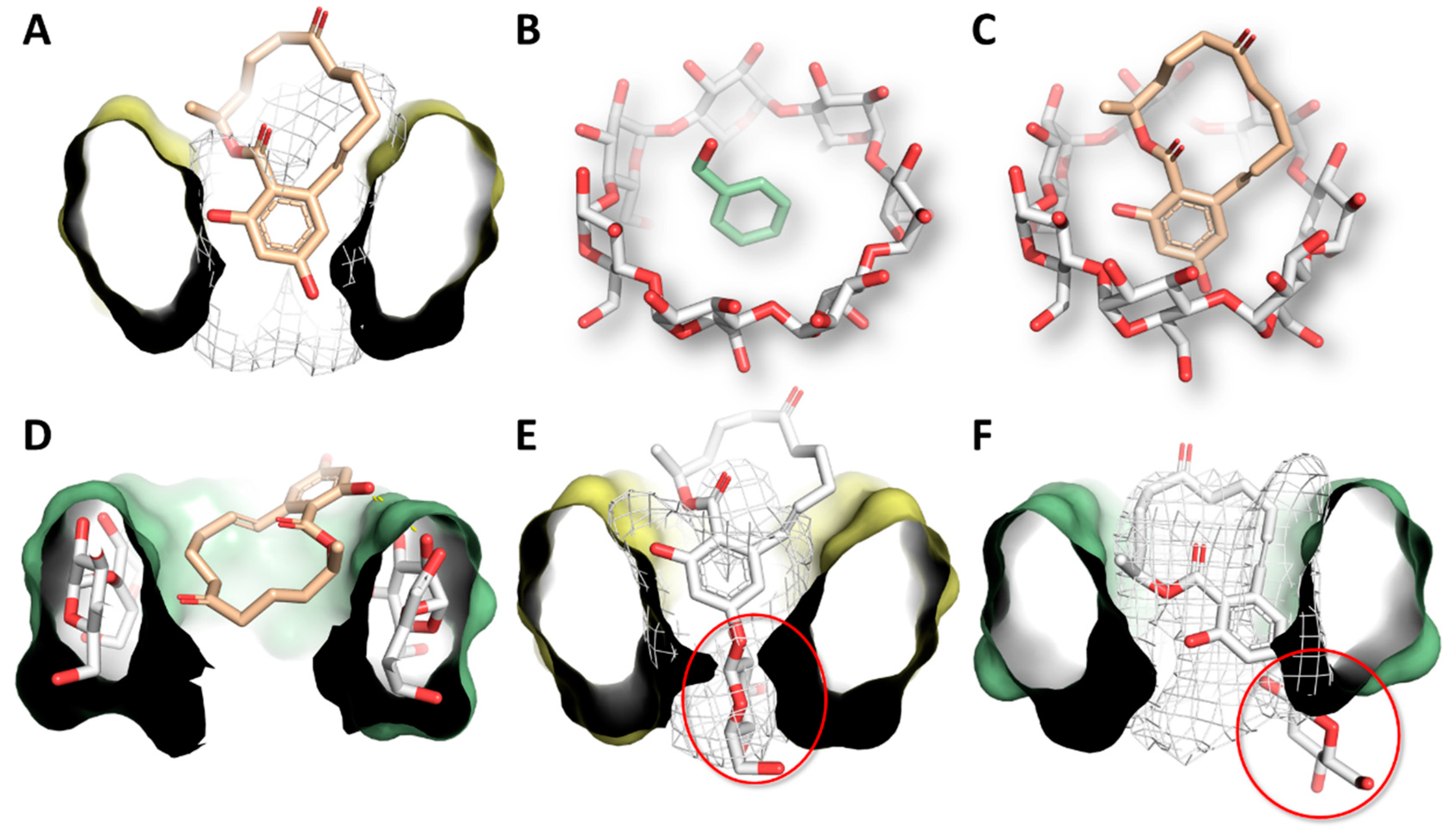
3.4. Molecular Modeling Studies
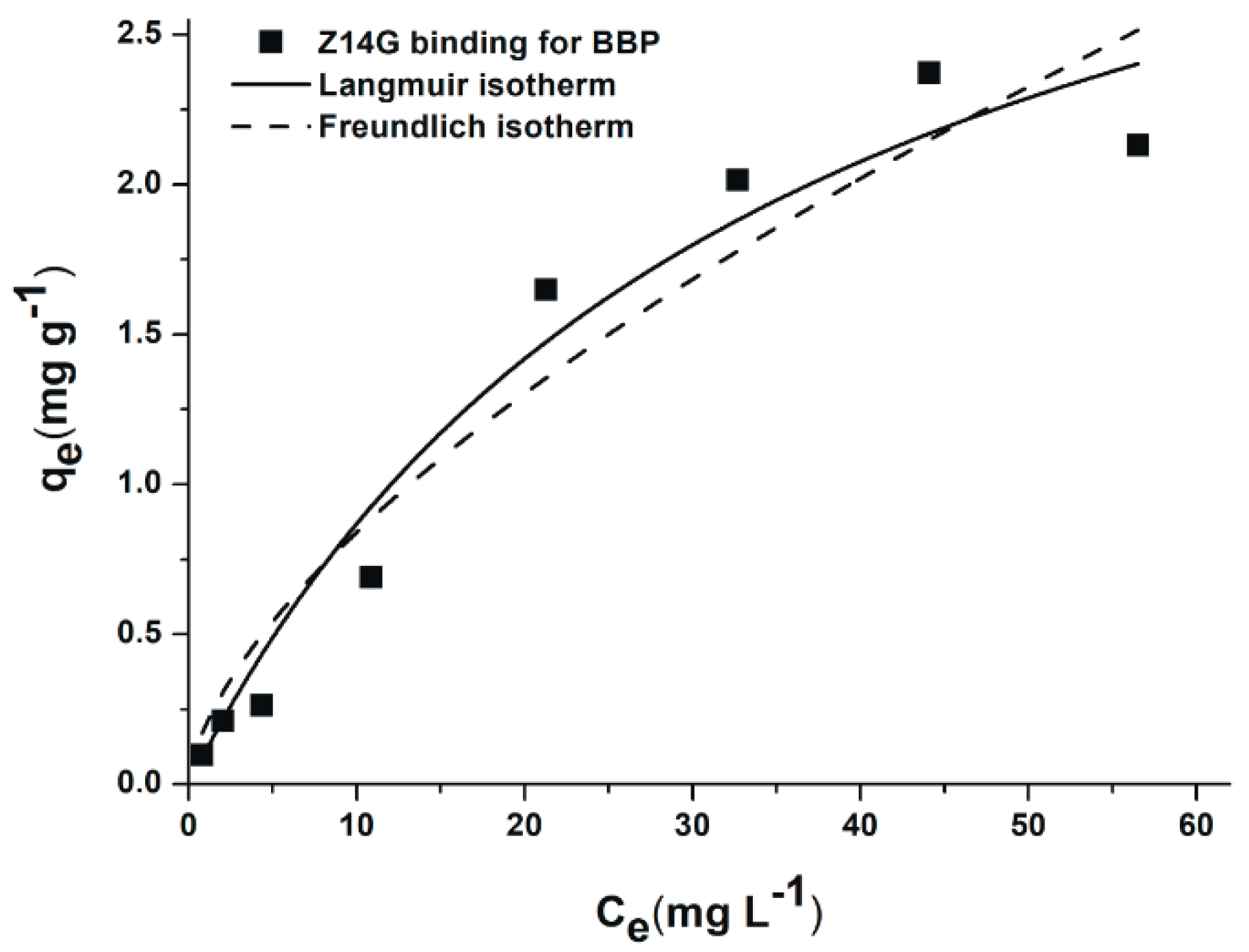
3.5. Extraction of Z14G from Aqueous Solution by BBP
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pitt, J.I. Mycotoxins. In Foodborne Infections and Intoxications, 4th ed.; Morris, J.G., Jr., Potter, M.E., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 30, pp. 409–418. [Google Scholar]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin. Microbiol. Rev. 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazar, S.; Omurtag, G.Z. Fumonisins, Trichothecenes and Zearalenone in Cereals. Int. J. Mol. Sci. 2008, 9, 2062–2090. [Google Scholar] [CrossRef] [PubMed]
- Maragos, C.M. Zearalenone occurrence and human exposure. World Mycotoxin J. 2010, 3, 369–383. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). Risks for animal health related to the presence of zearalenone and its modified forms in feed. EFSA J. 2017, 15, 4851. [Google Scholar] [CrossRef]
- Shier, W.; Shier, A.; Xie, W.; Mirocha, C. Structure-activity relationships for human estrogenic activity in zearalenone mycotoxins. Toxicon 2001, 39, 1435–1438. [Google Scholar] [CrossRef]
- Filannino, A.; Stout, T.A.; Gadella, B.M.; Sostaric, E.; Pizzi, F.; Colenbrander, B.; Dell’Aquila, M.E.; Minervini, F. Dose-response effects of estrogenic mycotoxins (zearalenone, alpha- and beta-zearalenol) on motility, hyperactivation and the acrosome reaction of stallion sperm. Reprod. Boil. Endocrinol. 2011, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Vianello, A.; Macri, F. Inhibition of plant cell membrane transport phenomena induced by zearalenone (F-2). Planta 1978, 143, 51–57. [Google Scholar] [CrossRef]
- Macrì, F.; Vianello, A. Zearalenone-dissipated Electrochemical Gradient in Higher Plant Mitochondria and Microsomes. J. Plant Physiol. 1990, 136, 754–757. [Google Scholar] [CrossRef]
- Berthiller, F.; Lemmens, M.; Werner, U.; Krska, R.; Hauser, M.-T.; Adam, G.; Schuhmacher, R. Short review: Metabolism of the Fusarium mycotoxins deoxynivalenol and zearalenone in plants. Mycotoxin Res. 2007, 23, 68–72. [Google Scholar] [CrossRef]
- Berthiller, F.; Crews, C.; Dall’Asta, C.; De Saeger, S.; Haesaert, G.; Karlovsky, P.; Oswald, I.P.; Seefelder, W.; Speijers, G.; Stroka, J. Masked mycotoxins: A review. Mol. Nutr. Food Res. 2013, 57, 165–186. [Google Scholar] [CrossRef]
- De Saeger, S.; Van Egmond, H.P. Special issue: Masked mycotoxins. World Mycotoxin J. 2012, 5, 203–206. [Google Scholar] [CrossRef]
- Zhang, Z.; Nie, D.; Fan, K.; Yang, J.; Guo, W.; Meng, J.; Zhao, Z.; Han, Z. A systematic review of plant-conjugated masked mycotoxins: Occurrence, toxicology, and metabolism. Crit. Rev. Food Sci. Nutr. 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Freire, L.; Sant’Ana, A.S. Modified mycotoxins: An updated review on their formation, detection, occurrence, and toxic effects. Food Chem. Toxicol. 2018, 111, 189–205. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Schuhmacher, R.; Adam, G.; Krska, R. Formation, determination and significance of masked and other conjugated mycotoxins. Anal. Bioanal. Chem. 2009, 395, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Rychlik, M.; Humpf, H.-U.; Marko, D.; Dänicke, S.; Mally, A.; Berthiller, F.; Klaffke, H.; Lorenz, N. Proposal of a comprehensive definition of modified and other forms of mycotoxins including “masked” mycotoxins. Mycotoxin Res. 2014, 30, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Poppenberger, B.; Berthiller, F.; Bachmann, H.; Lucyshyn, D.; Peterbauer, C.; Mitterbauer, R.; Schuhmacher, R.; Krska, R.; Glössl, J.; Adam, G. Heterologous Expression of Arabidopsis UDP-Glucosyltransferases in Saccharomyces cerevisiae for Production of Zearalenone-4-O-Glucoside. Appl. Environ. Microbiol. 2006, 72, 4404–4410. [Google Scholar] [CrossRef] [PubMed]
- Dall’Erta, A.; Cirlini, M.; Del Rio, D.; Galaverna, G.; Dall’Asta, C.; Dall’Asta, M.; Dall’Asta, M. Masked Mycotoxins Are Efficiently Hydrolyzed by Human Colonic Microbiota Releasing Their Aglycones. Chem. Res. Toxicol. 2013, 26, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Dellafiora, L.; Ruotolo, R.; Perotti, A.; Cirlini, M.; Galaverna, G.; Cozzini, P.; Buschini, A.; Dall’Asta, C. Molecular insights on xenoestrogenic potential of zearalenone-14-glucoside through a mixed in vitro/in silico approach. Food Chem. Toxicol. 2017, 108, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Szente, L. Highly soluble cyclodextrin derivatives: Chemistry, properties, and trends in development. Adv. Drug Deliv. Rev. 1999, 36, 17–28. [Google Scholar] [CrossRef]
- Crini, G. Review: A History of Cyclodextrins. Chem. Rev. 2014, 114, 10940–10975. [Google Scholar] [CrossRef] [PubMed]
- Szente, L.; Szemán, J. Cyclodextrins in Analytical Chemistry: Host–Guest Type Molecular Recognition. Anal. Chem. 2013, 85, 8024–8030. [Google Scholar] [CrossRef] [PubMed]
- Aghamohammadi, M.; Alizadeh, N. Fluorescence enhancement of the aflatoxin B1 by forming inclusion complexes with some cyclodextrins and molecular modeling study. J. Lumin. 2007, 127, 575–582. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, J.; Xu, H. Interactions of aflatoxin B1 and related secondary metabolites with native cyclodextrins and their potential utilization. Food Control. 2018, 94, 102–107. [Google Scholar] [CrossRef]
- Dall’Asta, C.; Faccini, A.; Galaverna, G.; Corradini, R.; Dossena, A.; Marchelli, R. Complexation of zearalenone and zearalenols with native and modified β-cyclodextrins. J. Incl. Phenom. Macrocycl. Chem. 2009, 64, 331–340. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, J.; Dong, L.; Lu, L.; Chen, F.; Hu, D.; Wang, X. A study of fluorescence properties of citrinin in β-cyclodextrin aqueous solution and different solvents. J. Lumin. 2012, 132, 1437–1445. [Google Scholar] [CrossRef]
- Poór, M.; Kunsági-Máté, S.; Sali, N.; Kőszegi, T.; Szente, L.; Peles-Lemli, B. Interactions of zearalenone with native and chemically modified cyclodextrins and their potential utilization. J. Photochem. Photobiol. B Boil. 2015, 151, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Poór, M.; Kunsági-Máté, S.; Szente, L.; Matisz, G.; Secenji, G.; Czibulya, Z.; Kőszegi, T. Interaction of ochratoxin A with quaternary ammonium beta-cyclodextrin. Food Chem. 2015, 172, 143–149. [Google Scholar] [CrossRef]
- Poór, M.; Matisz, G.; Kunsági-Máté, S.; Derdák, D.; Szente, L.; Lemli, B. Fluorescence spectroscopic investigation of the interaction of citrinin with native and chemically modified cyclodextrins. J. Lumin. 2016, 172, 23–28. [Google Scholar] [CrossRef]
- Maragos, C.M.; Appell, M.; Lippolis, V.; Visconti, A.; Catucci, L.; Pascale, M. Use of cyclodextrins as modifiers of fluorescence in the detection of mycotoxins. Food Addit. Contam. Part A 2008, 25, 164–171. [Google Scholar] [CrossRef]
- Dall’Asta, C.; Faccini, A.; Galaverna, G.; Corradini, R.; Dossena, A.; Marchelli, R. Complexation of the mycotoxin zearalenone with β-cyclodextrin: Study of the interaction and first promising applications. Mycotoxin Res. 2008, 24, 14–18. [Google Scholar] [CrossRef]
- Larionova, D.A.; Goryacheva, I.Y.; Van Peteghem, C.; De Saeger, S. Thin-layer chromatography of aflatoxins and zearalenones with β-cyclodextrins as mobile phase additives. World Mycotoxin J. 2011, 4, 113–117. [Google Scholar] [CrossRef]
- Poór, M.; Faisal, Z.; Zand, A.; Bencsik, T.; Lemli, B.; Kunsági-Máté, S.; Szente, L. Removal of Zearalenone and Zearalenols from Aqueous Solutions Using Insoluble Beta-Cyclodextrin Bead Polymer. Toxins 2018, 10, 216. [Google Scholar] [CrossRef] [PubMed]
- Appell, M.; Jackson, M.A. Synthesis and evaluation of cyclodextrin-based polymers for patulin extraction from aqueous solutions. J. Incl. Phenom. Macrocycl. Chem. 2010, 68, 117–122. [Google Scholar] [CrossRef]
- Appell, M.; Jackson, M.A. Sorption of Ochratoxin A from Aqueous Solutions Using β-Cyclodextrin-Polyurethane Polymer. Toxins 2012, 4, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellafiora, L.; Dall’Asta, C.; Cozzini, P. Ergot alkaloids: From witchcraft till in silico analysis. Multi-receptor analysis of ergotamine metabolites. Toxicol. Rep. 2015, 2, 535–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baroni, M.; Cruciani, G.; Sciabola, S.; Perruccio, F.; Mason, J.S. A Common Reference Framework for Analyzing/Comparing Proteins and Ligands. Fingerprints for Ligands And Proteins (FLAP): Theory and Application. J. Chem. Inf. Model. 2007, 47, 279–294. [Google Scholar] [CrossRef]
- Carosati, E.; Sciabola, S.; Cruciani, G. Hydrogen Bonding Interactions of Covalently Bonded Fluorine Atoms: From Crystallographic Data to a New Angular Function in the GRID Force Field. J. Med. Chem. 2004, 47, 5114–5125. [Google Scholar] [CrossRef]
- Dellafiora, L.; Warth, B.; Schmidt, V.; Del Favero, G.; Mikula, H.; Fröhlich, J.; Marko, D. An integrated in silico/in vitro approach to assess the xenoestrogenic potential of Alternaria mycotoxins and metabolites. Food Chem. 2018, 248, 253–261. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Boil. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Dellafiora, L.; Marchetti, M.; Spyrakis, F.; Orlandi, V.; Campanini, B.; Cruciani, G.; Cozzini, P.; Mozzarelli, A. Expanding the chemical space of human serine racemase inhibitors. Bioorg. Med. Chem. Lett. 2015, 25, 4297–4303. [Google Scholar] [CrossRef] [PubMed]
- Rollinger, J.M.; Schuster, D.; Baier, E.; Ellmerer, E.P.; Langer, T.; Stuppner, H. Taspine: Bioactivity-Guided Isolation and Molecular Ligand–Target Insight of a Potent Acetylcholinesterase Inhibitor from Magnolia x soulangiana. J. Nat. Prod. 2006, 69, 1341–1346. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, G.E.; Abraham, D.J. Hydrophobicity: Is LogPo/w more than the sum of its parts? Eur. J. Med. Chem. 2000, 35, 651–661. [Google Scholar] [CrossRef]
- Amadasi, A.; Dall’Asta, C.; Ingletto, G.; Pela, R.; Marchelli, R.; Cozzini, P. Explaining cyclodextrin–mycotoxin interactions using a ‘natural’ force field. Bioorg. Med. Chem. 2007, 15, 4585–4594. [Google Scholar] [CrossRef] [PubMed]
- Cozzini, P.; Ingletto, G.; Singh, R.; Dall’Asta, C. Mycotoxin Detection Plays “Cops and Robbers”: Cyclodextrin Chemosensors as Specialized Police? Int. J. Mol. Sci. 2008, 9, 2474–2494. [Google Scholar] [CrossRef] [PubMed]
- Cozzini, P.; Fornabaio, M.; Marabotti, A.; Abraham, D.J.; Kellogg, G.E.; Mozzarelli, A. Simple, Intuitive Calculations of Free Energy of Binding for Protein−Ligand Complexes. 1. Models without Explicit Constrained Water. J. Med. Chem. 2002, 45, 2469–2483. [Google Scholar] [CrossRef] [PubMed]
- Poór, M.; Zand, A.; Szente, L.; Lemli, B.; Kunsági-Máté, S. Interaction of α- and β-zearalenols with β-cyclodextrins. Molecules 2017, 22, 1910. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, M.L.; Berdini, V.; Hartshorn, M.J.; Mooij, W.T.M.; Murray, C.W.; Taylor, R.D.; Watson, P. Virtual Screening Using Protein−Ligand Docking: Avoiding Artificial Enrichment. J. Chem. Inf. Comput. Sci. 2004, 44, 793–806. [Google Scholar] [CrossRef]



| pH | Relative Increase in the Fluorescence of Z14G (I/I0 ± SEM) | |||||
|---|---|---|---|---|---|---|
| BCD | RAMEB | HPBCD | GCD | RAMEG | HPGCD | |
| 3.0 | 6.72 ± 0.12 | 6.23 ± 0.29 | 3.70 ± 0.09 | 5.03 ± 0.32 | 6.53 ± 0.08 | 5.38 ± 0.08 |
| 5.0 | 6.98 ± 0.19 | 6.16 ± 0.02 | 4.32 ± 0.03 | 5.49 ± 0.09 | 6.73 ± 0.22 | 5.79 ± 0.03 |
| 7.4 | 6.87 ± 0.10 | 5.93 ± 0.24 | 4.21 ± 0.21 | 4.92 ± 0.25 | 7.13 ± 0.10 | 5.53 ± 0.11 |
| 10.0 | 4.44 ± 0.14 | 4.99 ± 0.15 | 3.13 ± 0.14 | 3.62 ± 0.27 | 6.24 ± 0.17 | 4.37 ± 0.09 |
| pH | LogK (± SEM) | |||||
|---|---|---|---|---|---|---|
| BCD | RAMEB | HPBCD | GCD | RAMEG | HPGCD | |
| 3.0 | 2.74 ± 0.05 | 2.64 ± 0.07 | 2.55 ± 0.06 | 2.99 ± 0.06 | 3.18 ± 0.03 | 3.03 ± 0.02 |
| 5.0 | 2.81 ± 0.02 | 2.81 ± 0.05 | 2.71 ± 0.06 | 3.03 ± 0.05 | 3.27 ± 0.02 | 3.13 ± 0.01 |
| 7.4 | 2.93 ± 0.01 | 2.85 ± 0.05 | 2.82 ± 0.03 | 3.14 ± 0.02 | 3.25 ± 0.01 | 3.13 ± 0.03 |
| 10.0 | 2.10 ± 0.07 | 2.21 ± 0.05 | 1.95 ± 0.05 | 2.30 ± 0.05 | 2.38 ± 0.07 | 2.41 ± 0.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faisal, Z.; Fliszár-Nyúl, E.; Dellafiora, L.; Galaverna, G.; Dall’Asta, C.; Lemli, B.; Kunsági-Máté, S.; Szente, L.; Poór, M. Cyclodextrins Can Entrap Zearalenone-14-Glucoside: Interaction of the Masked Mycotoxin with Cyclodextrins and Cyclodextrin Bead Polymer. Biomolecules 2019, 9, 354. https://0-doi-org.brum.beds.ac.uk/10.3390/biom9080354
Faisal Z, Fliszár-Nyúl E, Dellafiora L, Galaverna G, Dall’Asta C, Lemli B, Kunsági-Máté S, Szente L, Poór M. Cyclodextrins Can Entrap Zearalenone-14-Glucoside: Interaction of the Masked Mycotoxin with Cyclodextrins and Cyclodextrin Bead Polymer. Biomolecules. 2019; 9(8):354. https://0-doi-org.brum.beds.ac.uk/10.3390/biom9080354
Chicago/Turabian StyleFaisal, Zelma, Eszter Fliszár-Nyúl, Luca Dellafiora, Gianni Galaverna, Chiara Dall’Asta, Beáta Lemli, Sándor Kunsági-Máté, Lajos Szente, and Miklós Poór. 2019. "Cyclodextrins Can Entrap Zearalenone-14-Glucoside: Interaction of the Masked Mycotoxin with Cyclodextrins and Cyclodextrin Bead Polymer" Biomolecules 9, no. 8: 354. https://0-doi-org.brum.beds.ac.uk/10.3390/biom9080354