Leveraging Container Technologies in a GIScience Project: A Perspective from Open Reproducible Research
Abstract
:1. Introduction
1.1. The SIOSE-INNOVA Project
1.2. Objectives
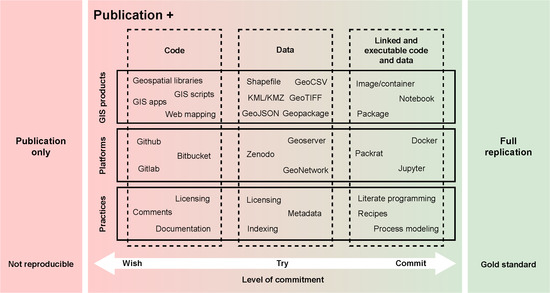
2. Open Reproducible Research in the Geospatial Domain
2.1. Collaborative Development and Code Sharing
- Python. This is the language with the greatest presence in the GIS realm. This is due to its use as a scripting language in the best-known GIS desktop applications, such as QGIS [38].
- R. This language is used for statistical computing, data visualisation, data science and geospatial analysis. It can also be used to access other GIS or inside GIS tools such as QGIS via connectors. The most relevant and useful R libraries are collected into the Spatial Task View (https://cran.r-project.org/view=Spatial).
- SQL. For many years, it has been used to query geospatial databases. SQL is at the heart of many GIS operations in spatial queries. It is used in software such as PostGIS [41], Spatialite and QGIS.
2.2. Geospatial Data Availability
- ESRI Shapefile [55]. This is a traditional and widely used vector data format. Normally, a Shapefile encloses at least three or four different files (.shp, .dbf, .shx and .prj) and defines a single type of geometry, which implies that each Shapefile can only store points, lines or polygons. This format, although widely used, was not intended as an open format when it was conceived. However, any GIScientist should know this format given the large amount of legacy data stored in this format and users that still use it.
- GeoCSV [56]. Comma-Separated Values (CSV) is a vector file format and is extensively used in any discipline. The CSV format allows work to be carried out using the most usual programs, such as Excel or Notepad. CSV is a plain text file where columns are separated using commas and rows by lines. More specifically, an optional geometry extension (GeoCSV) has been created that can store points (latitude and longitude) and Well-Known Text (WKT) standard geometries.
- GeoJSON [57]. This is an extension of JavaScript Object Notation (JSON) adding geospatial data. It offers the advantage of being a lightweight, easy-to-read notation for web applications. It is usually used as output for APIs and is a vector format.
- Spatialite/SQLite [58]. SpatiaLite is an open source library that extends the SQLite core with Spatial SQL capabilities. This is conceptually similar to the combination of PostgreSQL/PostGIS, but in this case, Spatialite is meant for embedded and portable geodatabases.
- NetCDF [61]. This is a data model for array-oriented scientific data that can also store both, vector and raster data. It is a recommended format to encode and distribute multi-dimensional and gridded geospatial data.
2.3. Linked Solutions in the Geospatial Context
3. A Brief Look at the Adoption of Containerisation in GISc Projects
- Selection of OSGeo tools. There are currently sixty-three different projects indexed on the OSGeo website, including OSGeo projects and community projects. Following the naming convention used on this website in determining featured tools that appears in the main menu, twenty-one tools have been selected from the seven categories defined in Table 1.
- Definition of searches. Searches were defined using the names of the OSGeo tools as query terms. Each search returned the list of images stored in Docker Hub, ordered by descending number of downloads (pulls). A filter was applied to set a maximum of 1000 results in order to ignore residual Docker images and build the analysis on those that are widely used.
- Execution of searches. Searches were executed by using the above-mentioned Docker Registry API. More specifically, we used a PowerShell module called PSDockerHub (https://github.com/beatcracker/PSDockerHub). The results were saved in a CSV file for each OSGeo project. From all the images, we obtained the name, description and number of downloads (pulls).
- Manual selection of results. After the search step, a manual phase is performed to detect possible Docker images not related to OSGeo tools. To reach this goal, the returned image results have been verified using the description field to check whether an OSGeo tool is used.
- Aggregation and final results. In the last step, an aggregation is performed on each tool to obtain a total number of downloads.
4. Enabling Research Reproducibility in the SIOSE-INNOVA Project
4.1. Benchmarking SQL and NoSQL Database Models
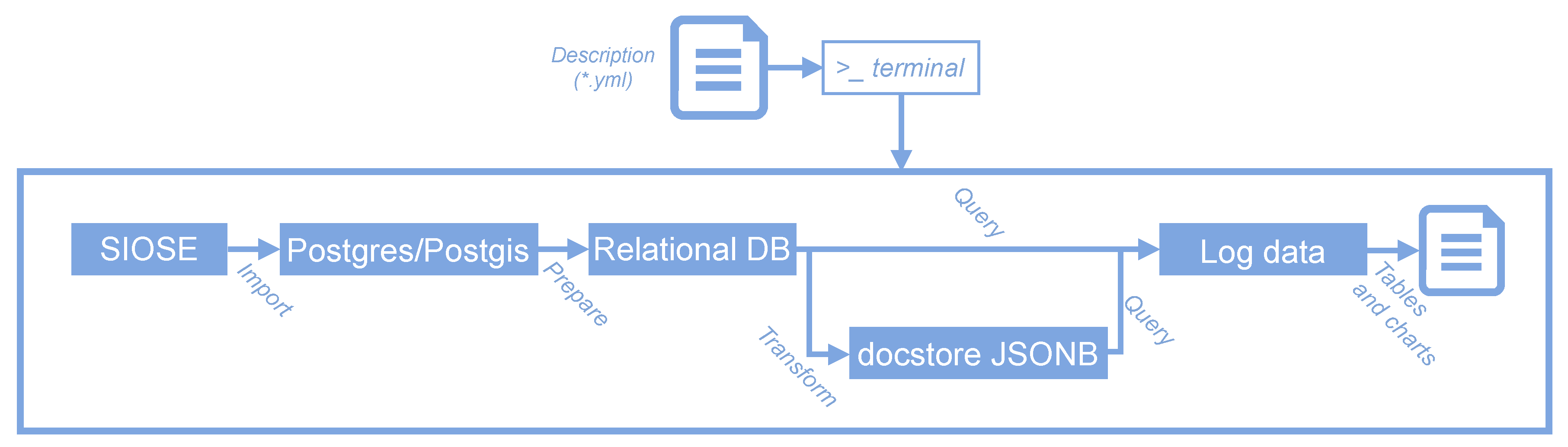
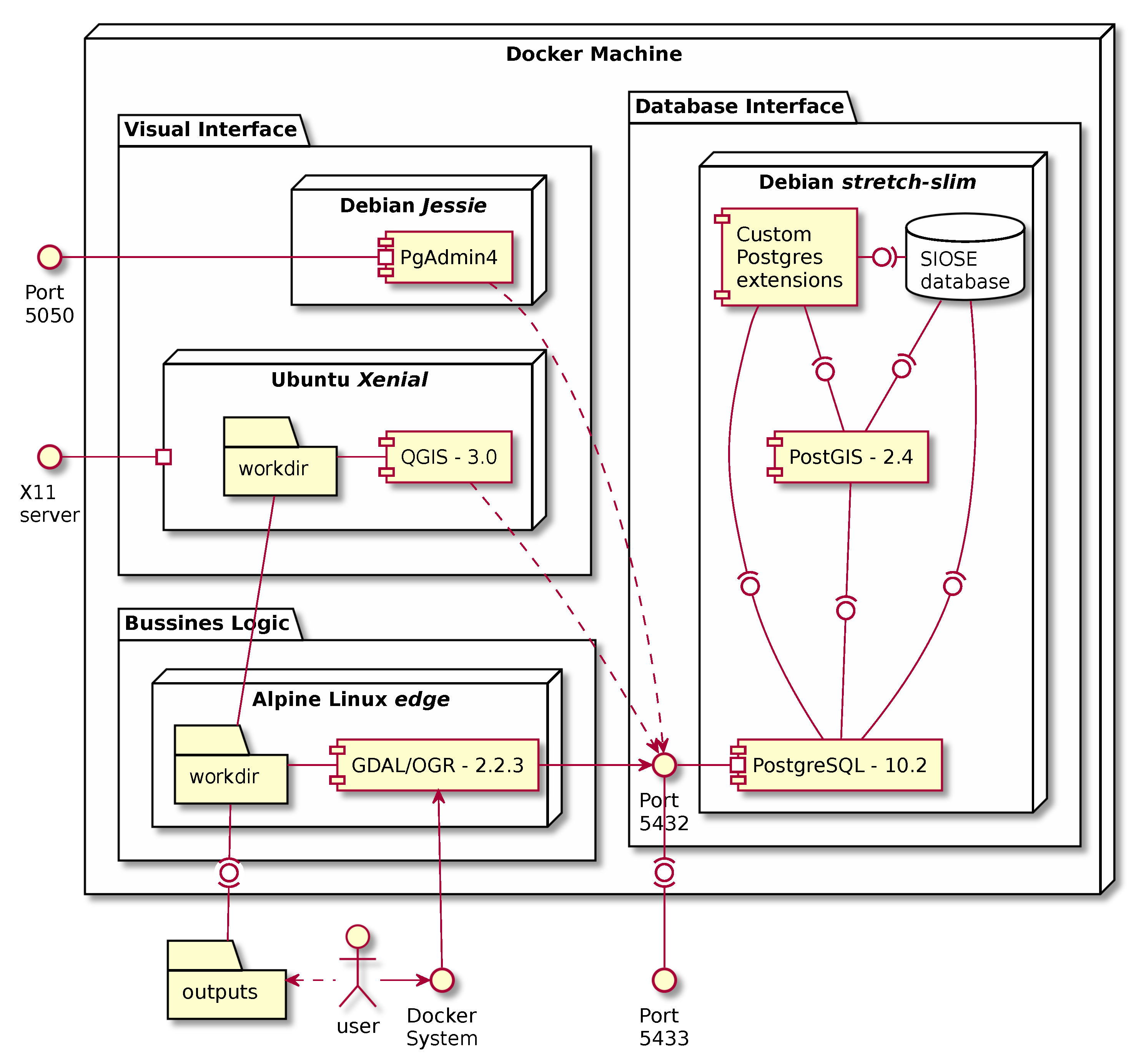
4.2. An ETL Process Using Containerisation
- The user launches the workflow using several docker run commands or one single execution of the deployment software (docker-compose up).
- Docker starts all the necessary services sequentially (see Figure 6).
- A PostgreSQL/PostGIS server is started which already contains a preloaded SIOSE database.
- SQL scripts are executed from the Makefile, creating spatial grids and statistics that will be used to split the database into GIS portable files. GNU Make will ensure that all files are created, and it will prevent any unnecessary overwrite operations from being carried out on any file.
- Then, the process loops over the cells of several spatial grids at different mapping scales.
- In every iteration, a spatial query is executed and intersects the SIOSE database with the corresponding area of interest (cell grid).
- Using the GDAL/OGR library, the selected SIOSE polygons together with a selection of LU/LC attributes are written into a new Geopackage file.
- This new Geopackage is compressed and stored in the working directory.
- Then the user can inspect the database or check the produced outputs using a containerised version of PGadmin4 or QGIS.
- When the user stops the process, Docker stops all services and frees all the unused computing resources.
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Meng, H.; Kommineni, R.; Pham, Q.; Gardner, R.; Malik, T.; Thain, D. An invariant framework for conducting reproducible computational science. J. Comput. Sci. 2015, 9, 137–142. [Google Scholar] [CrossRef]
- Barba, L.A. Terminologies for reproducible research. arXiv 2018, arXiv:1802.03311. [Google Scholar]
- Peng, R.D. Reproducible research in computational science. Science 2011, 334, 1226–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stodden, V. Reproducible research for scientific computing: Tools and strategies for changing the culture. Comput. Sci. Eng. 2012, 14, 13. [Google Scholar] [CrossRef] [Green Version]
- Brunsdon, C. Spatial science—Looking outward. Dialogues Hum. Geogr. 2014, 4, 45–49. [Google Scholar] [CrossRef] [Green Version]
- Gil, Y.; David, C.H.; Demir, I.; Essawy, B.T.; Fulweiler, R.W.; Goodall, J.L.; Karlstrom, L.; Lee, H.; Mills, H.J.; Oh, J.H.; et al. Toward the Geoscience Paper of the Future: Best practices for documenting and sharing research from data to software to provenance. Earth Space Sci. 2016, 3, 388–415. [Google Scholar] [CrossRef]
- Begley, C.G.; Ioannidis, J.P.A. Reproducibility in science: Improving the standard for basic and preclinical research. Circ. Res. 2015, 116, 116–126. [Google Scholar] [CrossRef] [Green Version]
- Baker, M. Is there a reproducibility crisis? Nature 2016, 533, 452–454. [Google Scholar] [CrossRef] [Green Version]
- Freedman, L.P.; Cockburn, I.M.; Simcoe, T.S. The economics of reproducibility in preclinical research. PLoS Biol. 2015, 13, 1–9. [Google Scholar] [CrossRef]
- Reinhart, C.M.; Rogoff, K.S. Growth in a time of debt. Am. Econ. Rev. 2010, 100, 573–578. [Google Scholar] [CrossRef] [Green Version]
- Herndon, T.; Ash, M.; Pollin, R. Does high public debt consistently stifle economic growth? A critique of Reinhart and Rogoff. Camb. J. Econ. 2014, 38, 257–279. [Google Scholar] [CrossRef] [Green Version]
- Open Science Collaboration. Estimating the reproducibility of psychological science. Science 2015, 349, aac4716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stodden, V. Implementing Reproducible Research; Chapman and Hall/CRC: Boca Raton, FL, USA, 2014; p. 440. [Google Scholar] [CrossRef]
- Singleton, A.D.; Spielman, S.; Brunsdon, C. Establishing a framework for Open Geographic Information science. Int. J. Geogr. Inf. Sci. 2016, 8816, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Goodchild, M.F. Geographical information science. Int. J. Geogr. Inf. Syst. 1992, 6, 31–45. [Google Scholar] [CrossRef]
- Ostermann, F.O.; Granell, C. Advancing Science with VGI: Reproducibility and Replicability of Recent Studies using VGI. Trans. GIS 2017, 21, 224–237. [Google Scholar] [CrossRef] [Green Version]
- Goodman, S.N.; Fanelli, D.; Ioannidis, J.P. What does research reproducibility mean? Sci. Transl. Med. 2016, 8, 341ps12. [Google Scholar] [CrossRef] [Green Version]
- Pontika, N.; Knoth, P.; Cancellieri, M.; Pearce, S. Fostering open science to research using a taxonomy and an eLearning portal. In Proceedings of the 15th International Conference on Knowledge Technologies and Data-Driven Business, Graz, Austria, 21–23 October 2015; p. 11. [Google Scholar]
- Antelman, K. Do Open Access Articles Have a Greater Research Impact? Coll. Res. Libr. 2004, 65. [Google Scholar] [CrossRef]
- Murray-Rust, P. Open data in science. Ser. Rev. 2008, 34, 52–64. [Google Scholar] [CrossRef]
- Perens, B. The open source definition. Open Sources Voices Open Source Revolut. 1999, 1, 171–188. [Google Scholar]
- CNIG Download Center. Available online: http://centrodedescargas.cnig.es. (accessed on 15 December 2019).
- Navarro-Carrión, J.T.; Zaragozí, B.; Ramón-Morte, A.; Valcárcel-Sanz, N. Should EU land use and land cover data be managed with a NOSQL document store? Int. J. Des. Nat. Ecodyn. 2016, 11, 438–446. [Google Scholar] [CrossRef] [Green Version]
- Giraud, T.; Lambert, N. Reproducible cartography. In International Cartographic Conference; Springer: Berlin/Heidelberg, Germany, 2017; pp. 173–183. [Google Scholar]
- Konkol, M.; Kray, C.; Pfeiffer, M. Computational reproducibility in geoscientific papers: Insights from a series of studies with geoscientists and a reproduction study. Int. J. Geogr. Inf. Sci. 2019, 33, 408–429. [Google Scholar] [CrossRef] [Green Version]
- Granell, C.; Nüst, D.; Ostermann, F.O.; Sileryte, R. Reproducible Research Is Like Riding a Bike; Technical Report; PeerJ Preprints: San Diego, CA, USA, 2018. [Google Scholar]
- Association of Geographic Information Laboratories in Europe (AGILE) Website. Available online: https://agile-online.org/ (accessed on 15 December 2019).
- Nüst, D.; Granell, C.; Hofer, B.; Konkol, M.; Ostermann, F.O.; Sileryte, R.; Cerutti, V. Reproducible research and GIScience: An evaluation using AGILE conference papers. PeerJ 2018, 6, e5072. [Google Scholar] [CrossRef]
- Skaggs, T.; Young, M.H.; Vrugt, J. Reproducible research in vadose zone sciences. Vadose Zone J. 2015, 14, vzj2015.06.0088. [Google Scholar] [CrossRef] [Green Version]
- Nüst, D.; Konkol, M.; Pebesma, E.; Kray, C.; Schutzeichel, M.; Przibytzin, H.; Lorenz, J. Opening the publication process with executable research compendia. D-Lib Mag. 2017, 23. [Google Scholar] [CrossRef]
- Barga, R.S.; Simmhan, Y.L.; Chinthaka, E.; Sahoo, S.S.; Jackson, J.; Araujo, N. Provenance for Scientific Workflows Towards Reproducible Research. IEEE Data Eng. Bull. 2010, 33, 50–58. [Google Scholar]
- BitBucket. Available online: https://www.bitbucket.com (accessed on 15 December 2019).
- GitLab. Available online: https://www.gitlab.com (accessed on 15 December 2019).
- Steiniger, S.; Hunter, A.J. The 2012 free and open source GIS software map - A guide to facilitate research, development, and adoption. Comput. Environ. Urban Syst. 2013, 39, 136–150. [Google Scholar] [CrossRef]
- Steiniger, S.; Bocher, E. An overview on current free and open source desktop GIS developments. Int. J. Geogr. Inf. Sci. 2009, 23, 1345–1370. [Google Scholar] [CrossRef]
- TIOBE Index TIOBE-The Software Quality Company. TIOBE Index| TIOBE- Softw. Qual. Co. [Electronic Resource]. 2018, Volume 1. Available online: https://www.Tiobe.Com/tiobe-Index/Access (accessed on 15 December 2019).
- OSGeo. Available online: http://www.osgeo.org (accessed on 15 December 2019).
- QGIS Development Team. QGIS geographic information system. Open Source Geospat. Found. Proj. Vers Ao 2015, 2. [Google Scholar]
- Hazzard, E. Openlayers 2.10 Beginner’s Guide; Packt Publishing Ltd: Birmingham, UK, 2011. [Google Scholar]
- Leaflet, J.S. Available online: https://leafletjs.com (accessed on 15 December 2019).
- PostGIS. Available online: https://postgis.net (accessed on 15 December 2019).
- Geoserver. Available online: http://geoserver.org (accessed on 15 December 2019).
- Geonetwork. Available online: http://geonetwork.org (accessed on 15 December 2019).
- Solutions, V. JTS Topology Suite: Technical Specifications. Version 2003, 1, 36. [Google Scholar]
- Neteler, M.; Bowman, M.H.; Landa, M.; Metz, M. GRASS GIS: A multi-purpose open source GIS. Environ. Model. Softw. 2012, 31, 124–130. [Google Scholar] [CrossRef] [Green Version]
- MapServer. Available online: http://www.saga-gis.org (accessed on 15 December 2019).
- GDAL. Available online: https://gdal.org. (accessed on 15 December 2019).
- Orfeo ToolBox. Available online: https://www.orfeo-toolbox.org. (accessed on 15 December 2019).
- Ram, K. Git can facilitate greater reproducibility and increased transparency in science. Source Code Biol. Med. 2013, 8, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Garderen, P. Archivematica: Using Micro-Services and Open-Source Software to Deliver a Comprehensive Digital Curation Solution. In Proceedings of the iPRES, Vienna, Austria, 19–24 September 2010. [Google Scholar]
- Faria, L.; Ferreira, M.; Castro, R.; Barbedo, F.; Henriques, C.; Corujo, L.; Ramalho, J.C. RODA: A service-oriented repository to preserve authentic digital objects. In Proceedings of the 4th International Conference on Open Repositories, Atlanta, GA, USA, 18–21 May 2009. [Google Scholar]
- Whyte, A.; Pryor, G. Open Science in Practice: Researcher Perspectives and Participation. IJDC 2011, 6, 199–213. [Google Scholar] [CrossRef]
- Benitez-Paez, F.; Comber, A.; Trilles, S.; Huerta, J. Creating a conceptual framework to improve the re-usability of open geographic data in cities. Trans. GIS 2018, 22, 806–822. [Google Scholar] [CrossRef] [Green Version]
- Stodden, V. The legal framework for reproducible scientific research: Licensing and copyright. Comput. Sci. Eng. 2008, 11, 35–40. [Google Scholar] [CrossRef] [Green Version]
- ESRI Shapefile Technical Description. Available online: https://www.esri.com/library/whitepapers/pdfs/shapefile.pdf (accessed on 15 December 2019).
- Stults, M.; Arko, R.; Davis, E.; Ertz, D.; Turner, M.; Trabant, C.; Valentine, D., Jr.; Ahern, T.; Carbotte, S.; Gurnis, M.; et al. GeoCSV: Tabular text formatting for geoscience data. AGU Fall Meet. Abstr. 2015. [Google Scholar]
- Butler, H.; Daly, M.; Doyle, A.; Gillies, S.; Hagen, S.; Schaub, T. The geojson format. Internet Eng. Task Force (IETF) 2016. [Google Scholar]
- SpatiaLite Webpage. Available online: https://www.gaia-gis.it/fossil/libspatialite/index (accessed on 15 December 2019).
- OGC GeoPackage. Available online: https://www.geopackage.org. (accessed on 15 December 2019).
- Open Geospatial Consortium. Available online: https://www.opengeospatial.org/standards. (accessed on 15 December 2019).
- netCDF. Available online: https://cdn.earthdata.nasa.gov/conduit/upload/497/ESDS-RFC-022v1.pdf. (accessed on 15 December 2019).
- Berners-Lee, T. Linked Data, 2006. Available online: https://www.w3.org/DesignIssues/LinkedData.html (accessed on 15 December 2019).
- Bizer, C.; Lehmann, J.; Kobilarov, G.; Auer, S.; Becker, C.; Cyganiak, R.; Hellmann, S. DBpedia-A crystallization point for the Web of Data. J. Web Semant. 2009, 7, 154–165. [Google Scholar] [CrossRef]
- Percivall, G. Progress in OGC web services interoperability development. In Standard-Based Data and Information Systems for Earth Observation; Springer: Berlin/Heidelberg, Germany, 2010; pp. 37–61. [Google Scholar]
- Giuliani, G.; Camara, G.; Killough, B.; Minchin, S. Earth Observation Open Science: Enhancing Reproducible Science Using Data Cubes, 2019. Available online: https://0-www-mdpi-com.brum.beds.ac.uk/2306-5729/4/4/147 (accessed on 15 December 2019).
- De la Beaujardiere, J. (Ed.) OpenGIS Web Map Service Implementation Specification, Version 1.3.0; Open Geospatial Consortium Inc.: Wayland, MA, USA, 2006; p. 85+. [Google Scholar]
- Vretanos, P.A. Web Feature Service Implementation Specification; Open Geospatial Consortium Specification: Wayland, MA, USA, 2005; pp. 04–094. [Google Scholar]
- Bröring, A.; Stasch, C.; Echterhoff, J. OGC Interface Standard 10-037: SOS 2.0 Interface Standard; Open Geospatial Consortium: Wayland, MA, USA, 2010. [Google Scholar]
- Nebert, D.; Whiteside, A. OpenGIS catalogue services specification (version 2.0). In OpenGIS Project Document 04-021r2; Open GIS Consortium Inc: Wayland, MA, USA, 2004. [Google Scholar]
- Schut, P. Opengis Web Processing Service Version 1.0.0; Technical report; Open Geospatial Consortium (OGC): Wayland, MA, USA, 2008. [Google Scholar]
- Nüst, D.; Stasch, C.; Pebesma, E. Connecting R to the sensor web. In Advancing Geoinformation Science for a Changing World; Springer: Berlin/Heidelberg, Germany, 2011; pp. 227–246. [Google Scholar]
- zenodo. Available online: https://www.zenodo.org (accessed on 15 December 2019).
- figshare. Available online: https://www.figshare.com (accessed on 15 December 2019).
- dspace. Available online: https://www.duraspace.org (accessed on 15 December 2019).
- ckan. Available online: https://ckan.org (accessed on 15 December 2019).
- Austin, C.C.; Brown, S.; Fong, N.; Humphrey, C.; Leahey, A.; Webster, P. Research data repositories: Review of current features, gap analysis, and recommendations for minimum requirements. IASSIST Q. 2016, 39, 24. [Google Scholar] [CrossRef]
- Creative Commons. Available online: https://creativecommons.org/licenses (accessed on 15 December 2019).
- Open Data Commons. Available online: https://opendatacommons.org/licenses (accessed on 15 December 2019).
- Benitez-Paez, F.; Degbelo, A.; Trilles, S.; Huerta, J. Roadblocks hindering the reuse of open geodata in Colombia and Spain: A data user’s perspective. ISPRS Int. J. Geo-Inf. 2018, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Trilles, S.; Díaz, L.; Huerta, J. Approach to facilitating geospatial data and metadata publication using a standard geoservice. ISPRS Int. J. Geo-Inf. 2017, 6, 126. [Google Scholar] [CrossRef] [Green Version]
- Mecklenburg, R. Managing Projects with GNU Make: The Power of GNU Make for Building Anything; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2004. [Google Scholar]
- Hutton, C.; Wagener, T.; Freer, J.; Han, D.; Duffy, C.; Arheimer, B. Most computational hydrology is not reproducible, so is it really science? Water Resour. Res. 2016, 52, 7548–7555. [Google Scholar] [CrossRef]
- Chirigati, F.; Rampin, R.; Shasha, D.; Freire, J. Reprozip: Computational reproducibility with ease. In Proceedings of the 2016 International Conference on Management of Data; ACM: New York, NY, USA, 2016; pp. 2085–2088. [Google Scholar]
- Stodden, V.; Miguez, S.; Seiler, J. Researchcompendia. org: Cyberinfrastructure for reproducibility and collaboration in computational science. Comput. Sci. Eng. 2015, 17, 12. [Google Scholar] [CrossRef]
- Howe, B. Virtual appliances, cloud computing, and reproducible research. Comput. Sci. Eng. 2012, 14, 36–41. [Google Scholar] [CrossRef]
- Dua, R.; Raja, A.R.; Kakadia, D. Virtualization vs containerization to support paas. In Proceedings of the 2014 IEEE International Conference on Cloud Engineering, Boston, MA, USA, 11–14 March 2014; pp. 610–614. [Google Scholar]
- Soltesz, S.; Pötzl, H.; Fiuczynski, M.E.; Bavier, A.; Peterson, L. Container-based operating system virtualization: A scalable, high-performance alternative to hypervisors. ACM SIGOPS Oper. Syst. Rev. 2007, 41, 275–287. [Google Scholar] [CrossRef]
- Turnbull, J. The Docker Book: Containerization Is the New Virtualization; Glitch: New York, NY, USA, 2014. [Google Scholar]
- Meadusani, S.R. Virtualization Using Docker Containers: For Reproducible Environments and Containerized Applications. Culminating Proj. Inf. Assur. 2018. [Google Scholar]
- Merkel, D. Docker: Lightweight linux containers for consistent development and deployment. Linux J. 2014, 2014, 2. [Google Scholar]
- Knoth, C.; Nüst, D. Reproducibility and practical adoption of geobia with open-source software in docker containers. Remote Sens. 2017, 9, 290. [Google Scholar] [CrossRef] [Green Version]
- Packrat—Introduction to renv. Available online: https://cran.r-project.org/web/packages/renv/vignettes/renv.html (accessed on 15 December 2019).
- Ushey, K. Renv—Introduction to renv. Available online: https://rstudio.github.io/renv/articles/renv.html (accessed on 15 December 2019).
- Nüst, D.; Hinz, M. containerit: Generating Dockerfiles for reproducible research with R. J. Open Source Softw. 2019, 4, 1603. [Google Scholar]
- Boettiger, C.; Eddelbuettel, D. An introduction to rocker: Docker containers for R. arXiv 2017, arXiv:1710.03675. [Google Scholar] [CrossRef] [Green Version]
- Knuth, D.E. Literate programming. Comput. J. 1984, 27, 97–111. [Google Scholar] [CrossRef] [Green Version]
- Kluyver, T.; Ragan-Kelley, B.; Pérez, F.; Granger, B.E.; Bussonnier, M.; Frederic, J.; Kelley, K.; Hamrick, J.B.; Grout, J.; Corlay, S.; et al. Jupyter Notebooks-a publishing format for reproducible computational workflows. In Proceedings of the ELPUB, Göttingen, Germany, 7–9 June 2016; pp. 87–90. [Google Scholar]
- Hillebrand, J.; Nierhoff, M.H. Mastering RStudio–Develop, Communicate, and Collaborate with R; Packt Publishing Ltd: Birmingham, UK, 2015. [Google Scholar]
- Trilles, S.; Granell, C.; Degbelo, A.; Bhattacharya, D. Interactive Guidelines: Public Communication of Data-based Research in Cities. PLoS ONE 2020. [Google Scholar] [CrossRef] [PubMed]
- p5js. Available online: https://p5js.org. (accessed on 15 December 2019).
- Trilles, S. OSGeo Tools in Docker Hub. 2019. Available online: https://www.google.com.hk/url?sa=t&rct=j&q=&esrc=s&source=web&cd=3&ved=2ahUKEwjehMyot4DoAhXZx4sBHeynDW4QFjACegQIAxAB&url=https%3A%2F%2Fwww.mdpi.com%2F2220-9964%2F9%2F3%2F138%2Fpdf&usg=AOvVaw1beLztM6wLMhEvqQ6d9NHs (accessed on 15 December 2019).
- Zaragozí, B.M.; Carrión, J.T.N. siose-innova/pg_siose_bench. 2020. Available online: https://www.google.com.hk/url?sa=t&rct=j&q=&esrc=s&source=web&cd=2&ved=2ahUKEwi03Jv2t4DoAhV7yYsBHZcLBowQFjABegQIAhAB&url=https%3A%2F%2Fwww.mdpi.com%2F2220-9964%2F9%2F3%2F138%2Fpdf&usg=AOvVaw1beLztM6wLMhEvqQ6d9NHs (accessed on 15 December 2019).



| Category | Tools |
|---|---|
| Content Management Systems | GeoNode |
| Desktop Applications | Marble, gvSIG Desktop, QGIS Desktop and GRASS GIS |
| Geospatial Libraries | GeoTools, Orfeo ToolBox, GDAL/OGR and GEOS |
| Metadata Catalogues | GeoNetwork and pycsw |
| Spatial Databases | PostGIS |
| Web Mapping | MapServer, MapFish, Deegree, OpenLayers, GeoMoose, Mapbender, PyWPS and GeoServer |
| Category | Tool | Downloads |
|---|---|---|
| Content Management Systems | GeoNode | 873,869 |
| Desktop Applications | GRASS GIS | 317 |
| gvSIG | 0 | |
| Marble | 0 | |
| QGIS | 392,882 | |
| Geospatial Libraries | GDAL/OGR | 1,653,481 |
| GEOS | 101,165 | |
| GeoTools | 2437 | |
| Orfeo Toolbox | 1736 | |
| Metadata Catalogues | GeoNetwork | 2,983,271 |
| pycsw | 482,912 | |
| Spatial Databases | PostGIS | 35,267,083 |
| Web Mapping | geomoose | 0 |
| Degree | 67 | |
| Geoserver | 1,731,538 | |
| MapBender | 986 | |
| MapFish | 0 | |
| MapServer | 1,170,538 | |
| OpenLayers | 320,234 | |
| pywps | 1162 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaragozí, B.M.; Trilles, S.; Navarro-Carrión, J.T. Leveraging Container Technologies in a GIScience Project: A Perspective from Open Reproducible Research. ISPRS Int. J. Geo-Inf. 2020, 9, 138. https://0-doi-org.brum.beds.ac.uk/10.3390/ijgi9030138
Zaragozí BM, Trilles S, Navarro-Carrión JT. Leveraging Container Technologies in a GIScience Project: A Perspective from Open Reproducible Research. ISPRS International Journal of Geo-Information. 2020; 9(3):138. https://0-doi-org.brum.beds.ac.uk/10.3390/ijgi9030138
Chicago/Turabian StyleZaragozí, Benito M., Sergio Trilles, and José T. Navarro-Carrión. 2020. "Leveraging Container Technologies in a GIScience Project: A Perspective from Open Reproducible Research" ISPRS International Journal of Geo-Information 9, no. 3: 138. https://0-doi-org.brum.beds.ac.uk/10.3390/ijgi9030138