1. Introduction
Nowadays, high resolution data encapsulating information regarding the vertical structure, such as elevation, surface, or canopy models derived from point clouds, have become more and more accessible. This calls for methods relying on 3D data and for suitable 3D approaches [
1] enabling the detection of fine-scale landscape or vegetation patterns [
2]. Such methods can complement state-of-the-art methods of vegetation classification [
3,
4] or can be used in a synergistic manner, such as the techniques of fusing LiDAR (Light Detection and Ranging) and multispectral imagery [
5,
6]. The 3D framework makes it possible to separate bare ground and canopy from LiDAR [
7] and to explore characteristics of the vertical structure of the canopy cover [
8] that were neglected for a long time. Thus, various canopy metrics related to the three-dimensional vegetation structure were brought to our attention: canopy height [
9] and crown dimensions [
10], the number of trees and tree heights [
11], the dendrometric parameters of species [
12], or comprehensive sets of forest stand structural parameters [
13]. Moreover, such canopy metrics were applied to the classification of the vegetation structure [
14]. Another application of the 3D approach was the automatic or semi-automatic detection of specific vegetation features [
15] regarded as structural elements of the three-dimensional vegetation cover [
16]. The methods based on the three-dimensional representation can be used in a complementary manner, since their performances depend on the type of forest under consideration [
17,
18].
There are other problems related to vegetation features that might be addressed in the 3D framework. They refer to important small-scale landscape elements [
19] such as shelterbelt trees, hedgerows, or scattered trees. The latter are prominent landscape features [
20] and represent a defining element of specific vegetation mosaics [
21]. Moreover, they play a key role in the provision of ecosystem services and therefore represent a vegetation feature that should be taken into account for future landscape design strategies [
22]. In this context, the identification of scattered trees from remote sensing data could be useful for foresters, managers, and landscape planners. On the other hand, by taking into account the scale at which they occur, a reliable approach could be based on high resolution data encompassing the vertical structure, such as high-resolution canopy height models [
23].
The identification of vegetation elements by using the 3D geospatial approach could be, in turn, linked to landscape ecological topics such as ecological connectivity, since isolated trees could contribute to a more functional connectivity [
24]. For instance, in vegetation mosaics such as wood-pasture landscapes, the scattered trees or the small forest patches provide food resources and facilitate the interconnectivity of the habitats for various species [
25,
26]. On the other hand, recent studies highlighted that for vegetation mosaics with a variable canopy height, the three-dimensional framework makes it possible to speak about the 3D ecological connectivity which augments the one relying on standard two-dimensional data, since it also refers to the structure of the vegetation strata [
27].
Many approaches to identifying landscape or vegetation features in 3D data rely on the use of statistical tools [
12,
13] or include a pixel-centered approach suitable for the two-dimensional set-up [
15]. Although very effective and efficient, such methods pay less attention to the 3D geometry of the objects of interest. We propose an approach that could enhance the existing ones by regarding the regular grids as discrete signals, reformulating the problem of identifying specific 3D patterns as a pattern recognition task and transferring approaches of this field or of related disciplines such as image or signal processing. For instance, multiresolution analysis is a technique for image processing, decomposition, and representation that analyzes the image data at different resolutions or scales without losing information [
28]. The signals can be considered in the discrete domain as sequences, or in the continuous domain as functions [
29]. The interplay between continuous and discrete is important, since through a discretization process, techniques stemming from the theory of continuous signals could be applied to the discrete ones. An example is provided by the wavelets, which are multi-resolution analysis tools that have emerged as an important tool for signal processing applications [
30]. Wavelet methods have been successfully used in the fine scale analysis of a wide range of ecological phenomena [
31,
32], including landscape pattern and heterogeneity studies [
33,
34,
35,
36], ecological modelling [
37,
38,
39], species ecology [
40,
41,
42], and environment analyses [
43,
44,
45,
46].
Wavelets come in various shapes; in consequence, each type is useful in analyzing particular data features. In ecological applications, the best known are the Haar wavelet, used for boundary detection [
47,
48], and the Mexican Hat wavelet. The latter represents a very appealing shape due to its resemblance to a tree, since it has a single central peak and symmetric lateral sides. In reality though, trees often have very disparate shapes and cannot perfectly match a symmetric pre-defined model. One workaround for addressing this issue is the possibility of varying the shape of the Mexican Hat wavelet by adapting the dilation coefficient that controls its geometry and the window size in order to better approximate the tree shape. In this context, the Mexican Hat wavelet has already been used for landscape feature extraction, such as trees [
39,
49,
50,
51] or pattern analysis [
35]. There is still a need to understand how the tree shapes and the geometry of the Mexican Hat can be linked when considering the vertical structure of the vegetation encapsulated in a surface or canopy model. For instance, one would need approaches that make this relationship independent of the use of alternative data, such as canopy models or surface models. Moreover, to the best of our knowledge, there is still a lack of a software tool compatible with standard GIS software that enables the automatic detection of pre-defined shapes, such as the Mexican Hat wavelet.
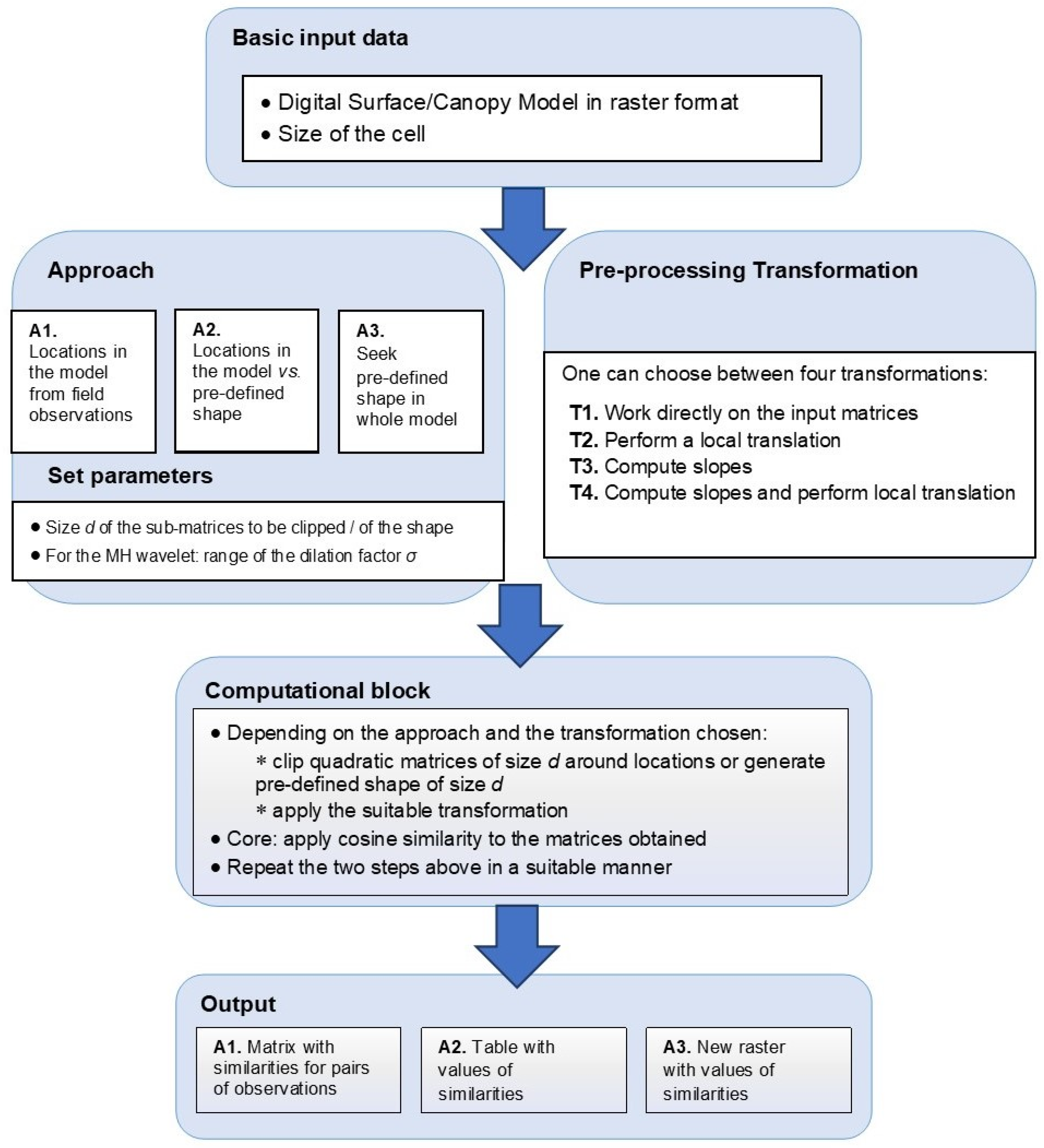
The analyses in this study were conducted under the assumption that shapes derived from high resolution digital surface/canopy height models can represent a proxy for vegetation patterns at a fine scale. The main research objective was to detect scattered trees by using the Mexican Hat wavelet as a geometric model. A subsidiary objective was to develop a software instrument compatible with standard GIS software, having as its main function the comparison between shapes or the detection of a pre-defined shape, such as the one described by the Mexican Hat wavelet or by other pre-defined geometric models.
3. Results
We now illustrate the use of the software tool in a study case, based on the data described in
Section 2.3. We considered the three approaches described in the methodology (approaches A1, A2, A3), thus combining data collected in situ with the use of pre-defined shapes, namely the Mexican Hat wavelets.
3.1. How Do the Pre-Processing Transformations Influence the Outcomes?
According to the study design described in
Section 2.5, we first computed the similarities for sub-matrices clipped around the field locations and applied a Principal Component Analysis for understanding the influence of various choices. The cumulated data variability explained by the first two components for
d = 3, 5, 7 was approximately 88.52%, 74.32%, and 73.77% respectively for Site 1 and 87.58%, 77.64%, and 75.23% respectively for Site 2 (see
Table 1 and
Table 2). On the other hand, except for one case, the percentage of the explained variability corresponding to the third component was less than the threshold value of 15.22% [
64]. The distances between the original objects and their projections in the plane generated by the first two components were well correlated. For Site 1, the values of the chosen goodness-of-fit measure were equal to 0.9877 (
d = 3), 0.9222 (
d = 5), and 0.9246 (
d = 7), and for Site 2, the values were equal to 0.9819 (
d = 3), 0.9543 (
d = 5), and 0.9603 (
d = 7). Therefore, one might conclude that the first two components are relevant for assessing the data variability, and that it is meaningful to interpret the original descriptors with respect to the new axes.
The values of the loadings for all the descriptors can be found in
Table 3 and
Table 4, and the corresponding biplots are represented in
Figure 4. Visually, the arrows corresponding to the transformation T3 are better aligned with the first principal axis. Indeed, in almost all cases (five out of six), DSM_T3 had the longest projection on the first component. This result hints that by applying the transformation T3 (namely, computing slopes towards the center), one better captures the variability of the vegetation features considered. We also notice that for
d = 5 and
d = 7, the arrow associated with transformation T4 is better correlated to the second principal component. Let us recall the interpretation of the transformations T3 and T4 and their relationship to the
z-values’ variation in first order and second order, which in turn could be related to indicators such as slopes and curvatures, respectively (e.g. [
65]). Thus, we might conclude that such indicators are relevant for explaining the variability of the tree shapes.
The direct use of the
z-values (i.e., the application of the identity transformation T1) may impact the computed similarity. This can be observed in
Figure 4, where T1 (relying directly on the
z-values) provides weak correlations for the values obtained when working on the CHM compared to the ones obtained for the DSM. The other methods have better correlations for the two data sources since, before computing the cosine similarity, a (local) transformation is performed, thus eliminating the impact of the absolute value.
In the following, we briefly describe a synthetic example, emphasizing the role played by the values compared to the effective shape of the matrices compared. We consider a 3 × 3 matrix representing a tree selected from Site 1, and we generate an arbitrary shape that hardly resembles a tree (e.g., the central value is a local minimum, while for a tree it is expected to be a local maximum).
.
However, the cosine similarity as obtained without performing any transformation is rather high (equal to 0.98). Let us notice that, by applying the transformations T2, T3, and T4, the values obtained are equal to 0.62, −0.87, and 0.57, respectively, and they clearly hint at a weak similarity between the tree and the synthetic shape.
In a nutshell, we concluded that the first step enabled us to perform comparisons between various transformations could be applied before computing the similarity. For the study case handled in the analyses, we found that working on the DSM and performing the transformation T3, corresponding to the z-values variation, better reflected the variability of the vegetation features.
3.2. To What Extent Do Trees Resemble Mexican Hat Wavelets?
According to the study design, we took into account the comparative analyses between the pre-processing transformations. We applied the transformation T3 before the comparison of the similarity values and chose the DSM as a reference model. The medians of the best matching values for Site 1 were equal to 0.97, 1.39, and 1.53 for the three selected cell sizes,
d = 3, 5, 7, respectively. For the second site, these values were 0.77, 0.47, and 0.26, respectively (see
Supplementary Material, Tables S1 and S2). In
Figure 5, we represented in box plots the quartiles corresponding to the best matching dilation factors for the two sites. Both from the median values and the graphical representation, one can deduce that there is a slight difference between the best matching dilation factors corresponding to the coniferous trees and the ones corresponding to the juniper shrubs, which could be due to the different 3D shapes of the vegetation features.
These outcomes could be put in a relationship with the geometry of the Mexican Hat wavelet (
Figure 1), as induced by the dilation factor
σ; for smaller values of
σ, the peak of the graph is higher, while the curve is sharper around the center and becomes quickly flat. This difference can be seen when comparing the behavior of the best matching values for the two sites when the size of the cell increases. Thus, for the site with coniferous trees, the median best matching value and the values defining the quartiles have a slight increase, while for the site with juniper shrubs, the opposite happens. This behavior could be explained by the different geometric shapes of the shrubs; the contour of the vegetation feature indicates a steep decrease around the center combined with flatness towards the borders of the clipped sub-matrix. On the other hand, shapes in nature are not necessarily regular, therefore artefacts might occur. For instance, some of the trees have an asymmetric shape due to natural variation or local environmental factors, are clumped together in small groups, or have intertwined canopies; this can be reflected by the
z-values in the grid. As shown in
Figure 6a on the CHM and
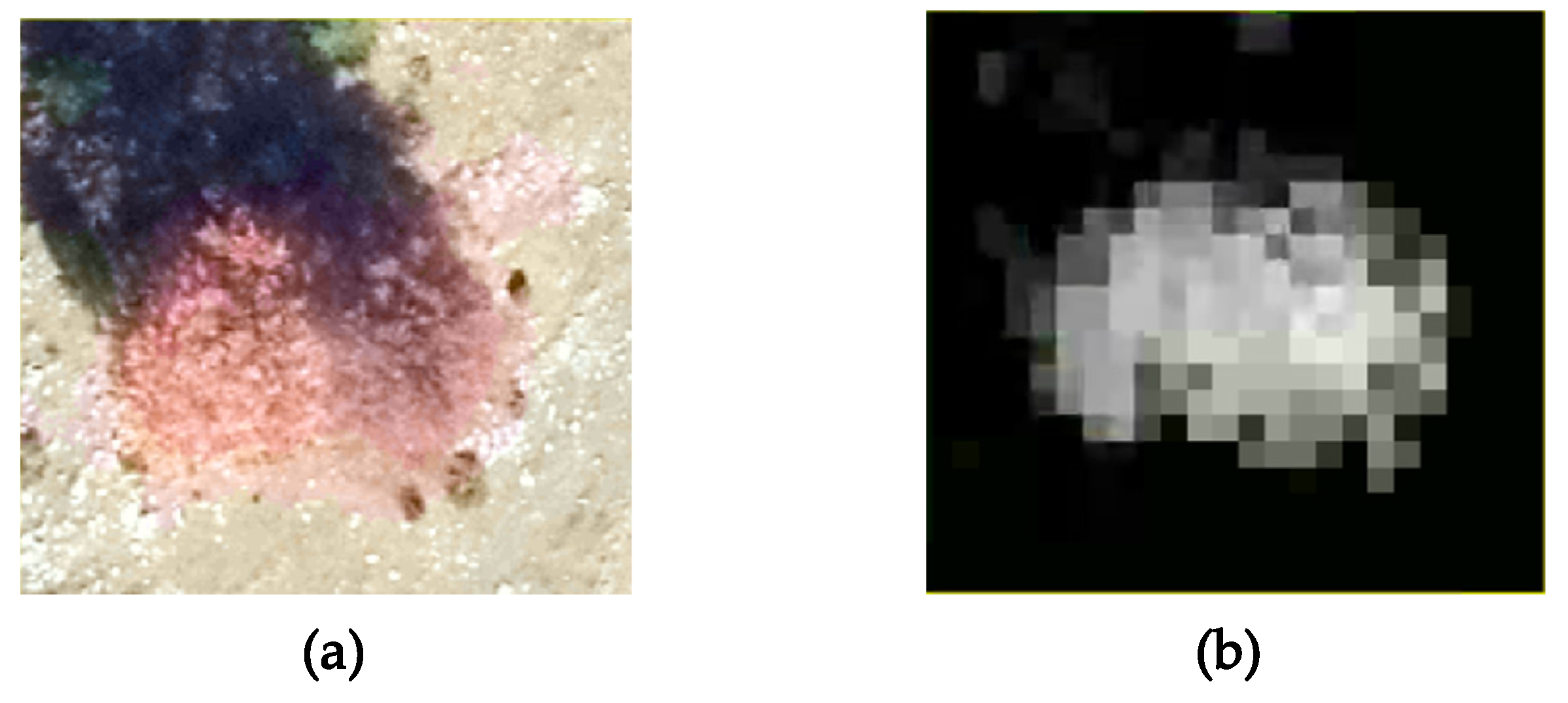
Figure 6b on the ortophotomap, some isolated trees can form small individual groups that present a pronounced asymmetry regarding their shape. Other than that, inherent errors due to the measurements or to the interpolation process that transformed the point cloud into a regular grid could lead to such asymmetries. Despite the irregularities, we might conclude that the distribution of the best matching dilation factors reflects the different vegetation features identified in the two sites.
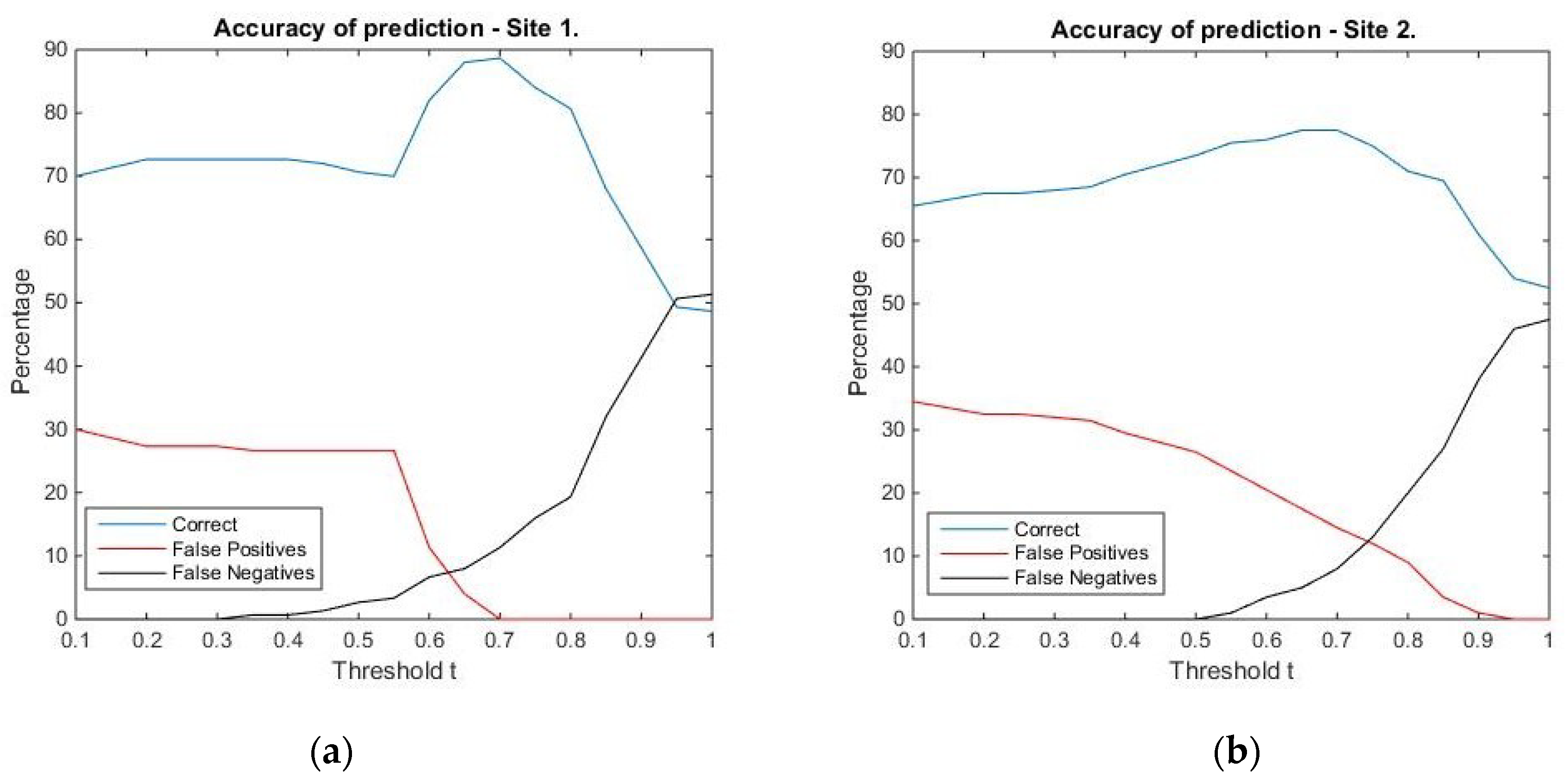
Threshold for similarity values corresponding to tree locations: The graphical representation (
Figure 7) hints that in the case of Site 1, the best prediction is obtained for the threshold value
t = 0.7, with an overall accuracy of 88.7%, an omission error of 11.3%, and no commission error. For Site 2, the best overall accuracy is equal to 77.5%, and it is obtained for the threshold values 0.65 and 0.7. We chose the former, since the percentage of true positives is 42.5%, compared to 39.5% for the latter. For this threshold value, the omission error is 5% and the commission error is 17.5%. The confusion matrices corresponding to the selected threshold values can be found in
Table 5 and
Table 6. Summing up, the threshold values 0.7 (for Site 1) and 0.65 (for Site 2) were chosen as reference values for comparing locations in the surface/canopy models with the pre-defined geometric shapes considered in the study.
3.3. How Can One Determine the Locations of Scattered Trees?
Several choices were necessary in the last stage, and they were determined by the outcomes of the previous steps. Thus, we chose the pre-processing transformation T3 (corresponding to the slopes) and the DSM as the data source, as indicated by the findings in
Section 3.1. The threshold value indicating a potential location of a tree was set at 0.7 for Sites 1 and R1, and it was set at 0.65 for Sites 2 and R2, as indicated by the analyses in
Section 3.2.
For the sites 1, 2, R1, and R2, we found 1079, 2723, 1107, and 5331 potential tree locations, respectively. Let us notice that, for Site 1, out of 77 trees identified in the field, 60 were indicated as potential tree locations, which yields an accuracy of 77.9%. For Site 2, out of 95 shrubs, 85 were found as potential locations (accuracy of 89.5%).
For visualization purposes, we transformed the matrices into rasters and then into shape files by using the suitable tools of the ArcGIS 10.1 software. An overlay with the orthophotomaps provided the spatial distribution of the potential locations (
Figure 8). One can notice that, visually, the outcomes are very plausible. For instance, in sites 1 and 2 most of them are situated in the middle part, namely in the pasture, where scattered trees can be indeed found in situ. In Site 1, few potential tree locations were indicated in the denser forests, hinting that the surface/canopy model around a scattered tree is different to the surface/canopy model around a tree situated in a forest. In Site R2, the trees belonging to the hedge situated in the northern half were identified as potential locations. The slight mismatch between the tree labels and the image could be due to the different spatial resolutions (the orthophotomap had a resolution of 12 cm, while the surface/canopy model had a resolution of 1 m).
In the
Supplementary Material, we included in
Figure S2 an application for two other sites with different characteristics. The outcomes are similar and confirm that the proposed approach could be successfully used in various environments.
4. Discussion
In summary, we might conclude that choosing suitable dilation factors for the Mexican Hat wavelet makes it possible to find potential locations for scattered trees or shrubs. Furthermore, as a subsequent step, one can extract from the canopy height model or the digital surface model an estimate of the tree heights. This is a general advantage of an approach relying on the vertical structure of the vegetation when compared to ones based only on two-dimensional imagery (even high-resolution).
Let us notice that, for Site 1, out of 77 trees identified in the field, 60 were indicated as potential tree locations, which yields an accuracy of 77.9%. For Site 2, out of 95 shrubs, 85 were found as potential locations (accuracy of 89.5%). Similar studies, which aimed to detect trees in remote sensing data, reported similar results. In [
17], six crown detection algorithms were investigated using aerial images under various forest conditions. The best accuracies reported varied from 60% for dense forest with a region growing algorithm to 99.7% for plantation with a marked point process algorithm. However, the authors warn that the algorithms are over-adapted for the specific forest type. Studies using data derived from LiDAR or Unmanned Aerial Vehicles (UAV) cite slightly higher accuracies ranging from 80% to 90% [
9,
18,
66,
67,
68,
69], while recent deep learning developments reached accuracies of >90% [
4,
70,
71].
LiDAR and other remote sensing data can be used to detect vegetation patterns at different scales, from small plants [
72,
73] to whole forests [
74]. It is important to note that the results are not immediately comparable between studies due to different data sets, pre-processing methods, environmental conditions (natural landscapes, urban areas, tree plantations), and not least, object detection techniques [
75]. Our method has the advantage that it is not bound to specific pre-processing tasks on LiDAR data. It works on any 2D matrix of elevation data, irrespective of the original source data, as we showed with the results on the CHM and DSM models. Also, unlike other tree detection methods in which model training is resource intensive and time consuming, the method we presented does not require such steps. Furthermore, the software tool contributes greatly to the reproducibility of the method, as it allows the user to easily follow the processing steps shown without the need to worry about implementing complex mathematical processing on matrix data. There is also a high degree of versatility to the tool and the described method, as it supports the detection of various shapes by simply using another wavelet filter or a generic 2D filter which describes the shape to be detected.
We illustrated the approach proposed and the functionalities of the software tool for solving a specific problem (the detection of scattered trees). However, the whole approach as well as the software instrument were conceived as a tool to be used in various analyses relying on regularly gridded surface/canopy models. Therefore, it offers the user the freedom to choose several approaches and parameters, providing appropriate outputs. Moreover, one could design, implement, and test alternative geometric shapes, depending on the problem under investigation. Thus, further possible applications refer to the comparison/detection of other vegetation patterns, geomorphologic features, or whole 3D landscape patterns.
5. Conclusions
The problem investigated in this paper was the detection of scattered trees in high resolution surface/canopy models. For achieving this aim, we developed and presented an approach and an accompanying software tool whose functionalities enable the comparison or the identification of landscape/vegetation patterns.
We proposed to address the issue by emphasizing the role of the 3D geometry of the objects of interest. Therefore, we explored the variability of regularly gridded digital surface/canopy models, regarded as discrete 2D signals. Thus, we adapted techniques stemming from pattern detection and we applied the cosine similarity measure to matrices whose entries were related to the vertical structure. We found that, when using surface/canopy models, the shape variability was better assessed by applying a pre-processing transformation based on slopes computation, corresponding to the variation in first order of the z-values.
In this paper, we presented an entire workflow that made it possible to find potential locations of scattered trees or shrubs from a digital surface model. This was achieved by using a pre-defined shape as a geometric model, the Mexican Hat wavelet resembling a coniferous tree. For two study sites, the trees and shrubs that were previously identified in the field were detected by our approach with accuracy rates of 77.9% and 89.5%. Moreover, in both sites and in four other sites with different characteristics chosen for validation, a visual check based on orthophotomaps confirmed the reliability of the outcomes.