2.2. Methodology
As mentioned earlier, GLMMs were developed in this study to model the spread of EAB. By using hierarchical layers in the analysis, GLMMs can be used to deal with instances where over-dispersion and correlation are evident. In the following discussion, the basic principles of the GLMM and its application to modelling the EAB spread are described.
Suppose there are n sampled points collected in a study area and divided into k groups based on spatial factors as
, and the total samples n is equal to
. GLMMs estimate the relationship between the mean value of the response variable
and risk predictors, which are connected by link function g(·) as shown in Equation (2),
where
and
. The linear predictor contains two different effects, namely fixed effects and random effects. All samples
among
k clusters share the same fixed effect based on the predictors
, which have coefficients denoted as
. The statistical inference on the parameter
shows the significance level of the predictors. The random effects denoted as
are identically distributed from a common density with mean zero
and covariance
. In Equation (2),
is an indication that the samples from the
ith cluster share the random effect
, which could be an intercept and random coefficient variables. In particular, samples
within the
ith cluster are modeled by the variable
, representing the random effect within their group. Hence, samples within different clusters are modeled by different random effects.
Since the mean values of the random effects are zero, each does not have an impact on the overall population mean. However, with different random effects, the linear predictors in Equation (2) could be different for the samples within different clusters, which can enhance model robustness and solve the earlier noted autocorrelation that is evident in the predictor data. To understand the random effects, consider the example of line fitting for clustered data, where the standard line fitting generates one line for all data points. However, when considering random effects, different lines could be generated to fit data points within different clusters. In this study, each observed point can be either a presence or absence point, which is modelled with the logistic link function. The observed samples from the same geographic neighborhood can be grouped into one spatial cluster and, therefore, different types of spatial random effects can be used in the proposed GLMMs.
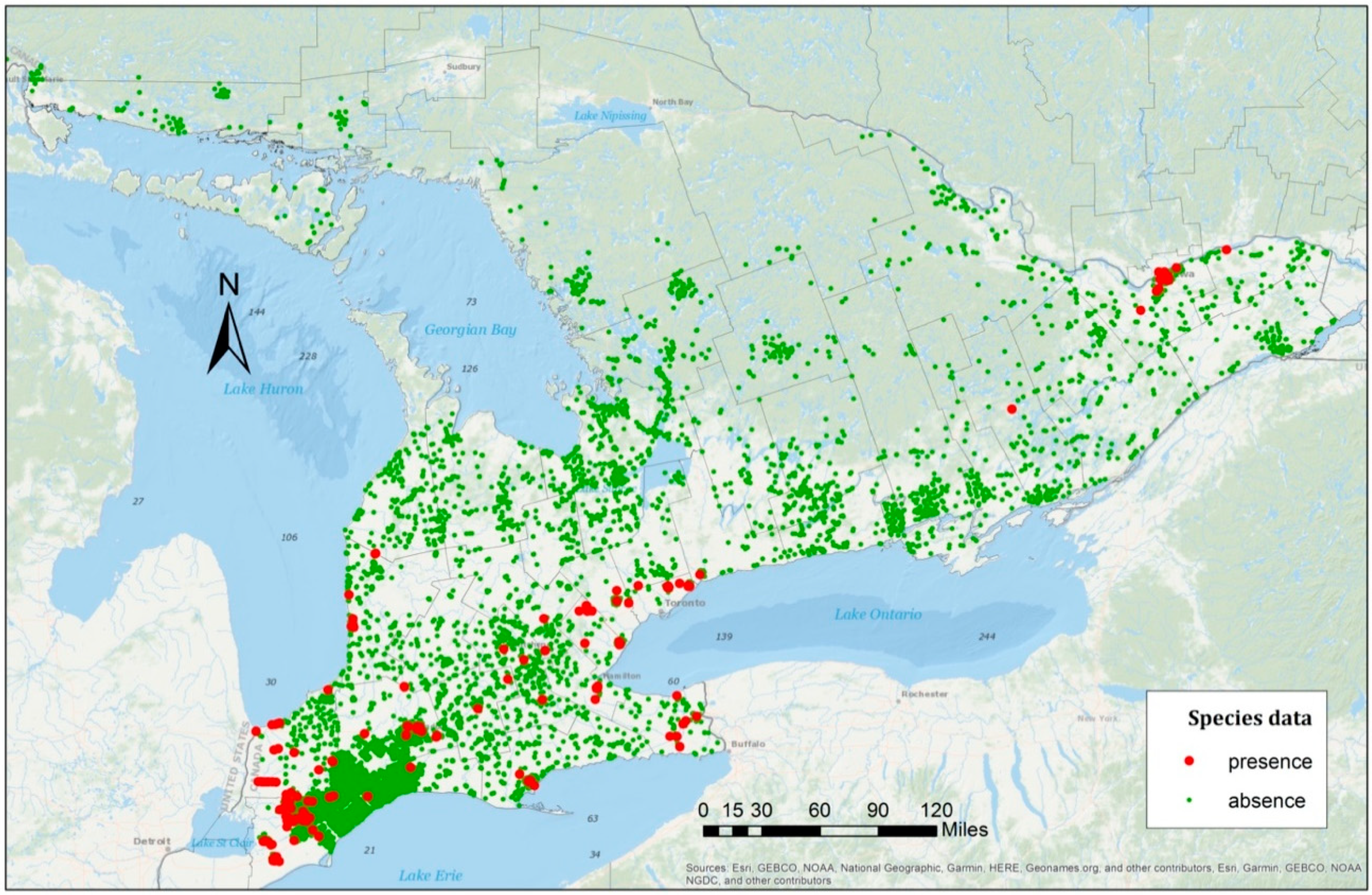
For the data used in this study, as shown in
Figure 2, clustered patterns from different counties were revealed and, thus, the samples were grouped by county in the first model. There are 46 counties in the species data, which can be represented by
, with
. Based on the collected data, some counties had many presence samples, while others were free from any observed EAB infestation. County boundaries are defined mainly for administrative purposes with no underlying environmental considerations. Hence, they can vary substantially in spatial extent and environmental conditions. Given this, a random effects model was also implemented, based on each sample’s geographic location. To accommodate this, we partitioned southern Ontario into 36 regions with an approximately equal size of 90 km by 150 km (
Figure 3).
Each region represented one spatial cluster and shared one random effect . Nine regions, namely R2, R3, R4, R5, R6, R30, R31, R35, and R36, had no surveyed data, hence these regions were not used in the random effects model. In comparison with the use of county random effects, the grid structure could be adjusted to accommodate different scenarios. For example, the clusters could be formed unevenly in their spatial structure based on local environmental features or other factors. Thus, two models were used to analyze the EAB distribution, including one GLMM with county random effects and another GLMM with regional random effects.
The estimate of regression coefficients
and random effects
can be obtained through numerical integration methods, and the predictive probabilities with the logistic link function calculated from
In the prediction model, the estimated parameters hold the asymptotic properties of consistency and normality. Thus, we can conduct statistical inference on each predictor with confidence. Meanwhile, values are the best linear unbiased predictions (BLUPs). A random intercept is used in the model for each cluster, which allows the cluster-specific effects to be differentiated. For example, in the clusters with more samples present, the prediction result may provide a more significant risk effect than those with lower risk.
In order to assess the significance of each predictor and overall performance of the proposed GLMMs, different statistical methods can be used. One approach is to examine the model fit by the residual deviance shown in Equation (4). This is a statistic measuring the difference of the estimated likelihoods
(the proposed model with the parameters of interest) and
(the saturated model, which can be overparametrized for each sample), namely,
which follows a chi-square distribution. This value can be used to conduct hypothesis testing to analyze the predictors in the model and compare models with different predictors. In addition, to select risk predictors with the best fit and control the model complexity, we can validate the model based on the Bayesian information criterion (BIC), namely,
This expression includes the maximum log-likelihood based on the candidate model in the subset s and the number of parameters k, which shows the level of complexity. The model with the lowest values of the Bayesian information criterion represents the best fit model. Other statistics, such as the Akaike information criterion, adjusted coefficient of determination (R2), and Cp statistic, can be used to compare different candidate models. Similar model selection results can be expected in most cases, while the BIC is a more restricted measure to deal with the overfit model for the large sample.
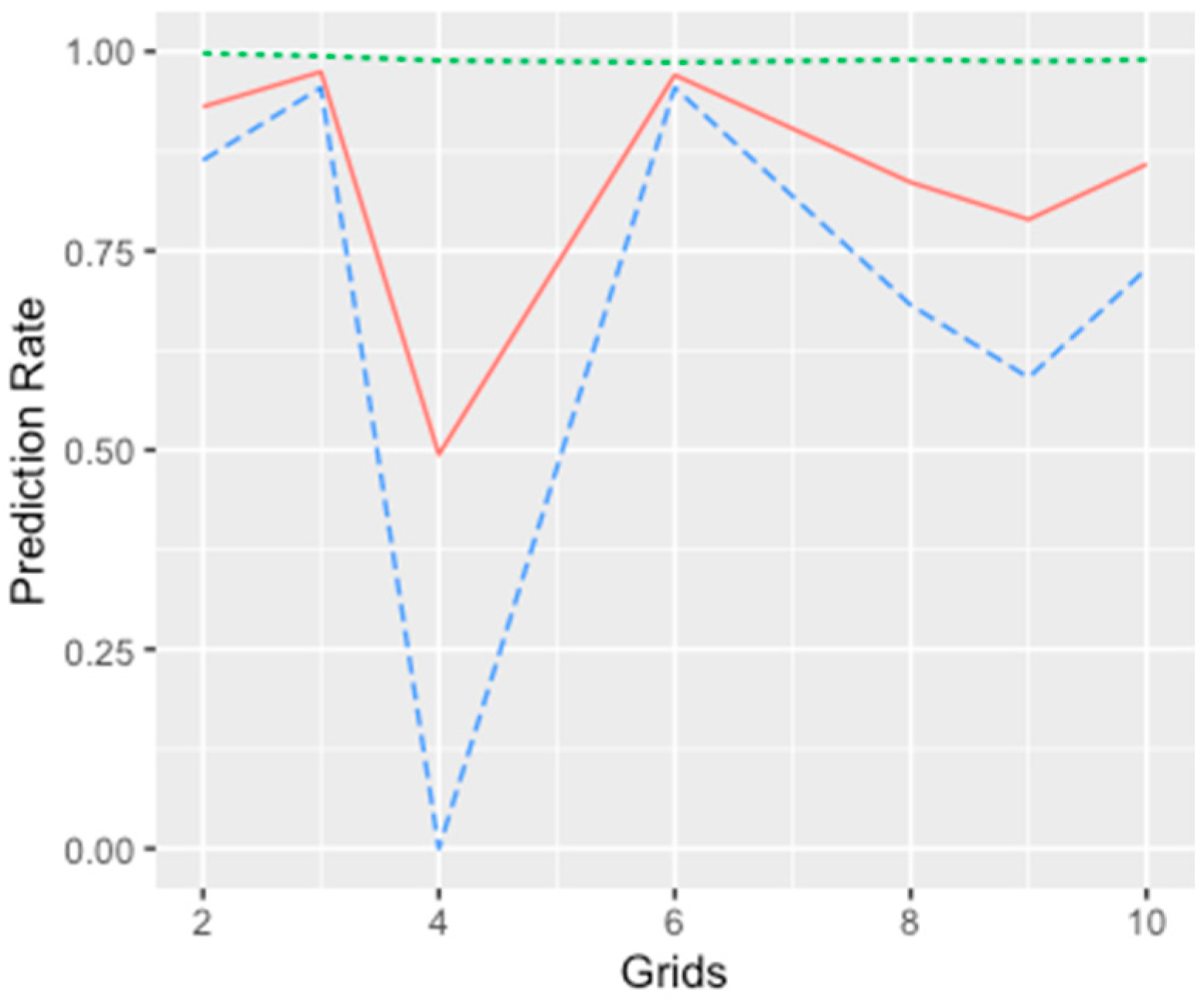
We conducted variable selection for the models using a stepwise process and estimated the values of the BIC with each step as the selection criterion. The order of adding a predictor at each step was based on each predictor’s significance level, and the BIC values were compared iteratively to determine which variable needed to be kept in the model. In addition, the prediction accuracy was another selection criterion proposed in the stepwise selection process. We applied a five-fold cross-validation with 100-fold replication to examine the predictive power of each model. Since the candidate models were proposed to analyze the spatial spread of the EAB, the training sets represented integrated information across all locations examined in the research. In each spatial cluster, we randomly sampled 80% of the presence–absence data and combined them as the training group to fit each model. The remaining data were applied to conduct validation for the classification accuracy.
For comparison, a logistic regression model, one of the most useful GLMs for binary responses (presence–absence data), was implemented. Since the model assumes independence between observations, the spatial autocorrelation present in the EAB data first needed to be removed. To do this, we measured the Euclidean distance between all sample locations based on ground coordinates and grouped the points through the use of clustering into 1000 neighborhoods. By randomly sampling one observation from each small neighborhood, a subset with 1000 samples was obtained. For this subset, the overall Moran’s I statistic was reduced to 0 with a large p-value, indicating that spatial autocorrelation was reduced to an insignificant level. Hence, the logistic regression model could be applied with confidence to the testing data for model comparison. The programming of the proposed model was conducted through the use of the R package ‘lme4’ function (
https://www.r-project.org/).