Mutations in SIX1 Associated with Branchio-oto-Renal Syndrome (BOR) Differentially Affect Otic Expression of Putative Target Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Obtaining Embryos and Microinjections
2.2. In Vitro Synthesis of mRNAs and Antisense RNA Probes
2.3. Histochemistry, In Situ Hybridization (ISH) and Analyses
2.4. QPCR Analyses
3. Results
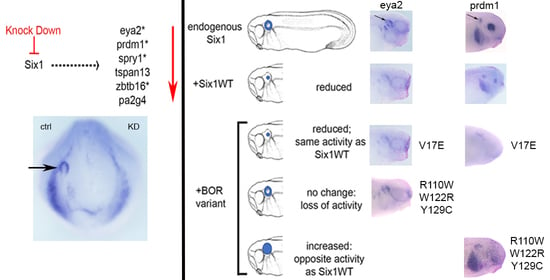
3.1. Expression of Several Otic Genes Requires Six1
3.2. BOR Mutations Differentially Reduce Cranial Expression of Putative Six1 Targets
3.3. BOR Mutations Differentially Affect Otic Gene Expression
4. Discussion
Do the Effects of BOR Variants Correlate with Patient Phenotypes?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fraser, F.C.; Sproule, J.R.; Halal, F. Frequency of the branchio-oto-renal (BOR) syndrome in children with profound hearing loss. Am. J. Med. Genet. 1980, 7, 341–349. [Google Scholar] [CrossRef]
- Kochhar, A.; Fischer, S.M.; Kimberling, W.J.; Smith, R.J. Branchio-oto-renal syndrome. Am. J. Med. Genet. A 2007, 143A, 1671–1678. [Google Scholar] [CrossRef]
- Smith, R.J. Branchiotorenal Spectrum Disorders. GeneReviews 2018. Available online: https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/books/NBK1380/ (accessed on 5 June 2021).
- Moody, S.A.; Neilson, K.M.; Kenyon, K.L.; Alfandari, D.; Pignoni, F. Using Xenopus to discover new genes involved in branchiootorenal spectrum disorders. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2015, 178, 16–24. [Google Scholar] [CrossRef] [Green Version]
- Pignoni, F.; Hu, B.; Zavitz, K.H.; Xiao, J.; Garrity, P.A.; Zipursky, S.L. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 1997, 91, 881–891. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, K.; Sato, S.; Ozaki, H.; Ikeda, K. Six family genes-structure and function as transcription factors and their roles in development. Bioessays 2000, 22, 616–626. [Google Scholar] [CrossRef]
- Kobayashi, M.; Nishikawa, K.; Suzuki, T.; Yamamoto, M. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev. Biol. 2001, 232, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, K.L.; Li, D.J.; Clouser, C.; Tran, S.; Pignoni, F. Fly SIX-type homeodomain proteins sine oculis and optix partner with different cofactors during eye development. Dev. Dyn. 2005, 234, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Ruf, R.G.; Berkman, J.; Wolf, M.T.; Nurnberg, P.; Gattas, M.; Ruf, E.M.; Hyland, V.; Kromberg, J.; Glass, I.; Macmillan, J.; et al. A gene locus for branchio-otic syndrome maps to chromosome 14q21.3-q24.3. J. Med. Genet. 2003, 40, 515–519. [Google Scholar] [CrossRef] [Green Version]
- Ruf, R.G.; Xu, P.X.; Silvius, D.; Otto, E.A.; Beekmann, F.; Muerb, U.T.; Kumar, S.; Neuhaus, T.J.; Kemper, M.J.; Raymond, R.M., Jr.; et al. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Natl. Acad. Sci. USA 2004, 101, 8090–8095. [Google Scholar] [CrossRef] [Green Version]
- Sanggaard, K.M.; Rendtorff, N.D.; Kjaer, K.W.; Eiberg, H.; Johnsen, T.; Gimsing, S.; Dyrmose, J.; Nielsen, K.O.; Lage, K.; Tranebjaerg, L. Branchio-oto-renal syndrome: Detection of EYA1 and SIX1 mutations in five out of six Danish families by combining linkage, MLPA and sequencing analyses. Eur. J. Hum. Genet. 2007, 15, 1121–1131. [Google Scholar] [CrossRef]
- Kochhar, A.; Orten, D.J.; Sorensen, J.L.; Fischer, S.M.; Cremers, C.W.; Kimberling, W.J.; Smith, R.J. SIX1 mutation screening in 247 branchio-oto-renal syndrome families: A recurrent missense mutation associated with BOR. Hum. Mutat. 2008, 29, 565. [Google Scholar] [CrossRef] [PubMed]
- Patrick, A.N.; Schiemann, B.J.; Yang, K.; Zhao, R.; Ford, H.L. Biochemical and functional characterization of six SIX1 Branchio-oto-renal syndrome mutations. J. Biol. Chem. 2009, 284, 20781–20790. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.M.; Krohn, P.; Baxi, A.P.; Tavares, A.L.P.; Sullivan, C.H.; Chillakuru, Y.R.; Majumdar, H.D.; Neilson, K.M.; Moody, S.A. Six1 proteins with human Branchio-oto-renal mutations differentially affect cranial gene expression and otic development. Dis. Models Mech. 2020, 13, dmm043489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlosser, G. Making senses development of vertebrate cranial placodes. Int. Rev. Cell Mol. Biol. 2010, 283, 129–234. [Google Scholar]
- Grocott, T.; Tambalo, M.; Streit, A. The peripheral sensory nervous system in the vertebrate head: A gene regulatory perspective. Dev. Biol. 2012, 370, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Saint-Jeannet, J.P.; Moody, S.A. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev. Biol. 2014, 389, 13–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moody, S.A.; LaMantia, A.-S. Transcriptional regulation of cranial sensory placode development. Curr. Top. Dev. Biol. 2015, 111, 301–350. [Google Scholar]
- Streit, A. Specification of sensory placode progenitors: Signals and transcription factor networks. Int. J. Dev. Biol. 2018, 62, 195–205. [Google Scholar] [CrossRef]
- Zheng, W.; Huang, L.; Wei, Z.B.; Silvius, D.; Tang, B.; Xu, P.X. The role of Six1 in mammalian auditory system development. Development 2003, 130, 3989–4000. [Google Scholar] [CrossRef] [Green Version]
- Brugmann, S.A.; Pandur, P.D.; Kenyon, K.L.; Pignoni, F.; Moody, S.A. Six1 promotes a placodal fate within the lateral neurogenic ectoderm by functioning as both a transcriptional activator and repressor. Development 2004, 131, 5871–5881. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, H.; Nakamura, K.; Funahashi, J.; Ikeda, K.; Yamada, G.; Tokano, H.; Okamura, H.O.; Kitamura, K.; Muto, S.; Kotaki, H.; et al. Six1 controls patterning of the mouse otic vesicle. Development 2004, 131, 551–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlosser, G.; Ahrens, K. Molecular anatomy of placode development in Xenopus laevis. Dev. Biol. 2004, 271, 439–466. [Google Scholar] [CrossRef]
- Zou, D.; Silvius, D.; Fritzsch, B.; Xu, P.X. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development 2004, 131, 5561–5572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bricaud, O.; Collazo, A. The transcription factor six1 inhibits neuronal and promotes hair cell fate in the developing zebrafish (Danio rerio) inner ear. J. Neurosci. 2006, 26, 10438–10451. [Google Scholar] [CrossRef]
- Konishi, Y.; Ikeda, K.; Iwakura, Y.; Kawakami, K. Six1 and Six4 promote survival of sensory neurons during early trigeminal gangliogenesis. Brain Res. 2006, 1116, 93–102. [Google Scholar] [CrossRef]
- Ikeda, K.; Ookawara, S.; Sato, S.; Ando, Z.; Kageyama, R.; Kawakami, K. Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev. Biol. 2007, 311, 53–68. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Kim, E.H.; Xu, P.X. Initiation of olfactory placode development and neurogenesis is blocked in mice lacking both Six1 and Six4. Dev. Biol. 2009, 326, 75–85. [Google Scholar] [CrossRef] [Green Version]
- Christophorou, N.A.; Bailey, A.P.; Hanson, S.; Streit, A. Activation of Six1 target genes is required for sensory placode formation. Dev. Biol. 2009, 336, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, K.; Kageyama, R.; Suzuki, Y.; Kawakami, K. Six1 is indispensable for production of functional progenitor cells during olfactory epithelial development. Int. J. Dev. Biol. 2010, 54, 1453–1464. [Google Scholar] [CrossRef] [PubMed]
- Bricaud, O.; Collazo, A. Balancing cell numbers during organogenesis: Six1a differentially affects neurons and sensory hair cells in the inner ear. Dev. Biol. 2011, 357, 191–201. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Xu, J.; Maire, P.; Xu, P.X. Six1 is essential for differentiation and patterning of the mammalian auditory sensory epithelium. PLoS Genet. 2017, 13, e1006967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, C.H.; Majumdar, H.D.; Neilson, K.M.; Moody, S.A. Six1 and Irx1 have reciprocal interactions during cranial placode and otic vesicle formation. Dev. Biol. 2019, 446, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Schlosser, G.; Awtry, T.; Brugmann, S.A.; Jensen, E.D.; Neilson, K.M.; Ruan, G.; Stammler, A.; Voelker, D.; Yan, B.; Zhang, C.; et al. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev. Biol. 2008, 320, 199–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maharana, S.K.; Schlosser, G. A gene regulatory network underlying the formation of pre-placodal ectoderm in Xenopus laevis. BMC Biol. 2018, 16, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, E.H.; Menezes, M.; Meyer, N.C.; Cucci, R.A.; Vervoort, V.S.; Schwartz, C.E.; Smith, R.J. Branchio-oto-renal syndrome: The mutation spectrum in EYA1 and its phenotypic consequences. Hum. Mutat. 2004, 23, 582–589. [Google Scholar] [CrossRef]
- Krug, P.; Morinière, V.; Marlin, S.; Koubi, V.; Gabriel, H.D.; Colin, E.; Bonneau, D.; Salomon, R.; Antignag, C.; Heidet, L. Mutation screening of the EYA1, SIX1, and SIX5 genes in a large cohort of patients harboring branchio-oto-renal syndrome calls into question the pathogenic role of SIX5 mutations. Hum. Mutat. 2011, 32, 183–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, Z.; Sato, S.; Ikeda, K.; Kawakami, K. Slc12a2 is a direct target of two closely related homeobox proteins, Six1 and Six4. FEBS J. 2005, 272, 3026–3041. [Google Scholar] [CrossRef]
- Jusiak, B.; Karandikar, U.C.; Kwak, S.J.; Wang, F.; Wang, H.; Chen, R.; Mardon, G. Regulation of Drosophila eye development by the transcription factor Sine oculis. PLoS ONE 2014, 9, e89695. [Google Scholar] [CrossRef] [Green Version]
- Yan, B.; Neilson, K.M.; Ranganathan, R.; Streit, A.; Moody, S.A. Microarray identification of novel genes downstream of Six1, a critical factor in cranial placode, somite and kidney development. Dev. Dyn. 2015, 244, 181–210. [Google Scholar] [CrossRef] [Green Version]
- Riddiford, N.; Schlosser, G. Dissecting the pre-placodal transcriptome to reveal presumptive direct targets of Six1 and Eya1 in cranial placodes. eLife 2016, 5, e17666. [Google Scholar] [CrossRef]
- Li, J.; Zhang, T.; Ramakrishnan, A.; Fritzsch, B.; Xu, J.; Wong, E.Y.M.; Loh, Y.E.; Ding, J.; Shen, L.; Xu, P.X. Dynamic changes in cis-regulatory occupancy by Six1 and its cooperative interactions with distinct cofactors drive lineage-specific gene expression programs during progressive differentiation of the auditory sensory epithelium. Nucleic Acids Res. 2020, 48, 2880–2896. [Google Scholar] [CrossRef]
- Neilson, K.M.; Abbruzzesse, G.; Kenyon, K.; Bartolo, V.; Krohn, P.; Alfandari, D.; Moody, S.A. Pa2G4 is a novel Six1 co-factor that is required for neural crest and otic development. Dev. Biol. 2017, 421, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, T.; Sato, S.; Ikeda, K.; Yajima, H.; Kawakami, K. Multiple evolutionarily conserved enhancers control expression of Eya1. Dev. Dyn. 2008, 237, 3142–3156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xu, J.; Xu, P. Eya2 expression during mouse embryonic development revealed by Eya2lacZ knockin reporter and homozygous mice show mild hearing loss. Dev. Dyn. 2021, 14. [Google Scholar] [CrossRef]
- Wilm, T.P.; Solnica-Krezel, L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development 2005, 132, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Powell, D.R.; Hernandez-Lagunas, L.; LaMonica, K.; Artinger, K.B. Prdm1a directly activates foxd3 and tfap2a during zebrafish neural crest specification. Development 2013, 140, 3445–3455. [Google Scholar] [CrossRef] [Green Version]
- Eguchi, R.; Yoshigai, E.; Koga, T.; Kuhara, S.; Tashiro, K. Spatiotemporal expression of prdm genes during Xenopus development. Cytotech 2015, 67, 711–719. [Google Scholar] [CrossRef] [Green Version]
- Prajapati, R.S.; Hintze, M.; Streit, A. PRDM1 controls the sequential activation of neural, neural crest and sensory progenitor determinants. Development 2019, 146. [Google Scholar] [CrossRef]
- Yang, X.; Kilgallen, S.; Andreeva, V.; Spicer, D.B.; Pinz, I.; Friesel, R. Conditional expression of Spry1 in neural crest causes craniofacial and cardiac defects. BMC Dev. Biol. 2010, 10, 48. [Google Scholar] [CrossRef] [Green Version]
- Simrick, S.; Lickert, H.; Basson, M.A. Sprouty genes are essential for the normal development of epibranchial ganglia in the mouse embryo. Dev. Biol. 2011, 358, 147–155. [Google Scholar] [CrossRef] [Green Version]
- Wright, K.; Rogers, A.; Zhang, J.; Shim, K. Cooperative and independent functions of FGF and Wnt signaling during early inner ear development organogenesis. BMC Dev. Biol. 2015, 15, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Termini, C.M.; Gillette, J.M. Tetraspanins function as regulators of cellular signaling. Front. Cell Dev. Biol. 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felthaus, O.; Gosau, M.; Morsczeck, C. ZBTB16 induces osteogenic differentiation marker genes in dental follicle cells independnet from RUNX2. J. Periodontol. 2014, 85, e144–e151. [Google Scholar] [CrossRef]
- Yan, F.; Jia, P.; Yoshioka, H.; Suzuki, A.; Iwata, J.; Zhao, Z. A developmental stage-specific network approach for studying dynamic co-regulation of transcription factors and microRNAs during craniofacial development. Development 2020, 147, dev192948. [Google Scholar] [CrossRef] [PubMed]
- Neilson, K.M.; Pignoni, F.; Yan, B.; Moody, S. A Developmental expression patterns of candidate cofactors for vertebrate six family transcription factors. Dev. Dyn. 2010, 239, 3446–3466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, P.A.; Fu, H.; Luo, P.; Zhao, Q.; Yu, J.; Ferrari, A.; Tenzen, T.; Yuk, D.; Tsung, E.F.; Cai, Z.; et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 2004, 306, 2255–2257. [Google Scholar] [CrossRef]
- Ko, H.R.; Chang, Y.S.; Park, W.S.; Ahn, J. Opposing roles of the two isoforms of ErbB3 binding protein 1 in human cancer cells. Int. J. Cancer 2016, 139, 1202–1208. [Google Scholar] [CrossRef] [Green Version]
- Moody, S.A. Lineage tracing and fate mapping. Cold Spring Harb. Protoc. 2018. [Google Scholar] [CrossRef]
- Klein, S.L. The first cleavage furrow demarcates the dorsal-ventral axis in Xenopus embryos. Dev. Biol. 1987, 120, 299–304. [Google Scholar] [CrossRef]
- Miyata, S.; Kageura, H.; Kihara, H.K. Regional differences of proteins in isolated cells of early embryos of Xenopus laevis. Cell Differ. 1987, 21, 47–52. [Google Scholar] [CrossRef]
- Moody, S.A.; Kline, M.J. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat. Embryol. 1990, 182, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Nieuwkoop, P.D.; Faber, J. Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis; Garland Publishing: New York, NY, USA, 1994. [Google Scholar]
- Yan, B.; Neilson, K.M.; Moody, S.A. foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev. Biol. 2009, 329, 80–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohto, H.; Kamada, S.; Tago, K.; Tominaga, S.I.; Ozaki, H.; Sato, S.; Kawakami, K. Cooperation of six and eya in activation of their target genes through nuclear translocation of eya. Mol. Cell Biol. 1999, 19, 6815–6824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Oghi, K.A.; Zhang, J.; Krones, A.; Bush, K.T.; Glass, C.K.; Nigam, S.K.; Aggarwal, A.K.; Mass, R.; Rose, D.W.; et al. Eya protein phosphatase activity regulates Six1-dach-eya transcriptional effects in mammalian organogenesis. Nature 2003, 426, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.J.; Davies, E.L.; Doyon, L.; Rebay, I. Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell Biol. 2003, 23, 5989–5999. [Google Scholar] [CrossRef] [Green Version]
- Peshkin, L.; Lukyanov, A.; Kalocsay, M.; Gage, R.M.; Wang, D.Z.; Pells, T.J.; Karimi, K.; Vize, P.D.; Wuhr, M.; Kitschner, M.W. The protein repertoire in early vertebrate embryogenesis. bioRXiv 2019. [Google Scholar] [CrossRef]
- Neilson, K.M.; Keer, S.; Bousquet, N.; Majumdar, H.D.; Kenyon, K.L.; Alfandari, D.; Moody, S.A. Mcrs1 interacts with Six1 to influence early craniofacial and otic development. Dev. Biol. 2020, 467, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Manaligod, J.M.; Weeks, D.L. EYA1 mutations associated with the branchio-oto-renal syndrome result in defective otic development in Xenopus laevis. Biol. Cell 2010, 102, 277–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoskins, B.E.; Cramer, C.H.; Silvius, D.; Zou, D.; Raymond, R.M.; Orten, D.J.; Kimberling, W.J.; Smith, R.J.; Weil, D.; Petit, C.; et al. Transcription factor SIX5 is mutated in patients with branchio-oto-renal syndrome. Am. J. Hum. Genet. 2007, 80, 800–804. [Google Scholar] [CrossRef] [Green Version]
- Patrick, A.N.; Cabrera, J.H.; Smith, A.L.; Chen, X.S.; Ford, H.L.; Zhao, R. Structure-fucntion analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome. Nat. Struct. Mol. Biol. 2013, 20, 447–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kribelbauer, J.F.; Rastogi, C.; Bussemaker, H.J.; Mann, R.S. Low-affinity binding sites and the transcription factor specificity paradox in eukaryotes. Annu. Rev. Cell Dev. Biol. 2019, 35, 357–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Gene | Forward Sequence | Reverse Sequence |
|---|---|---|
| eya2 | CCTCGGACGACAATGGACAA | CAGTCAACTCCCCCATGGAC |
| prdm1 | AAGGAACACGGTTTGGACCA | TGAAGTGCTGGAAGTCACCA |
| spry1 | TGCTTGCACAGAGGTTTTCAG | TTGTAGCTCCATCTGTAGTGATCT |
| tspan13 | CATGCGCGTCTCTGGCTATA | AGCCCCACAGGTGTCATTTT |
| zbtb16 | GGGTGTGAGCTCTGTGGAAA | ACACACAAATGCCTTTGCCC |
| pa2g4 | GCCTGAAAATGAAAACCTCCC | TTCCACAACTCCCATTCTCG |
| Gene | Is Six1 Required? | Does Six1 Activate or Repress? | V17E | R110W | W122R | Y129C |
|---|---|---|---|---|---|---|
| eya2 * | yes | repress | same as Six1WT | low: loss of activity high: same as Six1WT | low: loss of activity high: same as Six1WT | low: loss of activity high: same as Six1WT |
| prdm1 * | yes | repress | more repressive than Six1WT | low: loss of activity high: loss of activity & opposite activity | low: loss of activity high: loss of activity & opposite activity | low: loss of activity high: loss of activity & opposite activity |
| spry1 * | yes | both | loss of activity | low: loss of activity high: same as Six1WT | low: loss of activity high: loss of activity | low: same as Six1WT high: loss of activity |
| tspan13 | yes | low: repress high: activate | same as Six1WT | low: loss of activity high: same as Six1WT | low: loss of activity high: same as Six1WT | low: loss of activity high: same as Six1WT |
| zbtb16 * | yes | --- | same as Six1WT | low: loss of activity high: loss of activity | low: loss of activity high: loss of activity | low: loss of activity high: loss of activity |
| pa2g4 | yes | low: activate high: repress | same as Six1WT | low: loss of activity high: loss of activity | low: loss of activity high: loss of activity | low: same as Six1WT high: loss of activity |
| Patients | hearing loss hyoid fistulae preauricular pits [12] | hearing loss hyoid cysts (v) preauricular pits (v) pinnae defect (v) renal (v) [10,12] | hearing loss hyoid fistulae (v) preauricular pits (v) [11] | hearing loss hyoid fistulae (v) preauricular pits (v) [9] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehdizadeh, T.; Majumdar, H.D.; Ahsan, S.; Tavares, A.L.P.; Moody, S.A. Mutations in SIX1 Associated with Branchio-oto-Renal Syndrome (BOR) Differentially Affect Otic Expression of Putative Target Genes. J. Dev. Biol. 2021, 9, 25. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb9030025
Mehdizadeh T, Majumdar HD, Ahsan S, Tavares ALP, Moody SA. Mutations in SIX1 Associated with Branchio-oto-Renal Syndrome (BOR) Differentially Affect Otic Expression of Putative Target Genes. Journal of Developmental Biology. 2021; 9(3):25. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb9030025
Chicago/Turabian StyleMehdizadeh, Tanya, Himani D. Majumdar, Sarah Ahsan, Andre L. P. Tavares, and Sally A. Moody. 2021. "Mutations in SIX1 Associated with Branchio-oto-Renal Syndrome (BOR) Differentially Affect Otic Expression of Putative Target Genes" Journal of Developmental Biology 9, no. 3: 25. https://0-doi-org.brum.beds.ac.uk/10.3390/jdb9030025