Binding Properties of Photosynthetic Herbicides with the QB Site of the D1 Protein in Plant Photosystem II: A Combined Functional and Molecular Docking Study
Abstract
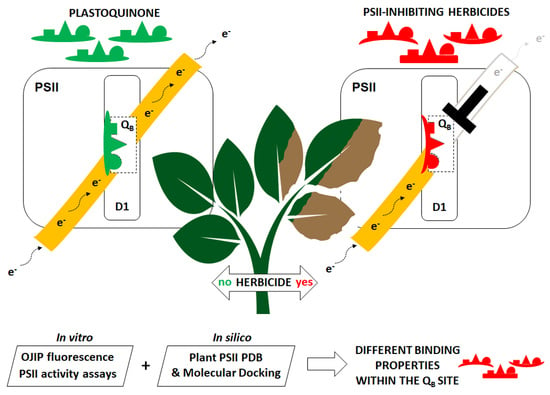
:1. Introduction
2. Results and Discussion
2.1. Structural Conservation of the D1 Protein and QB Binding Site among Oxygenic Photosynthetic Organisms
2.2. Estimation of the Herbicide Binding Affinity for the QB Binding Site of the D1 Protein by Photochemical and Fluorescence Assays of Photosystem II Inhibition
2.3. Molecular Docking Study of the Interaction of Herbicides in the QB Binding Site of the D1 Protein
3. Conclusions
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Thylakoid Isolation
4.3. Thylakoid Treatment with Herbicides
4.4. Photochemical Assay
4.5. Fluorescence Assay
4.6. Sequence Alignment
4.7. Molecular Docking Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Shen, J.R. The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 2015, 66, 23–48. [Google Scholar] [CrossRef] [Green Version]
- Nelson, N.; Junge, W. Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 2015, 84, 659–683. [Google Scholar] [CrossRef]
- Zouni, A.; Witt, H.T.; Kern, J.; Fromme, P.; Krauss, N.; Saenger, W.; Orth, P. Crystal structure of photosystem II from Synechococcus elongatus at 3.8 Å resolution. Nature 2001, 409, 739–743. [Google Scholar] [CrossRef]
- Umena, Y.; Kawakami, K.; Shen, J.-R.; Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 2011, 473, 55–60. [Google Scholar] [CrossRef]
- Tanaka, A.; Fukushima, Y.; Kamiya, N. Two different structures of the oxygen-evolving complex in the same polypeptide frameworks of photosystem II. J. Am. Chem. Soc. 2017, 139, 1718–1721. [Google Scholar] [CrossRef]
- Ago, H.; Adachi, H.; Umena, Y.; Tashiro, T.; Kawakami, K.; Tian, N.K.L.; Han, G.; Kuang, T.; Liu, Z.; Wang, F.; et al. Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 2016, 291, 5676–5687. [Google Scholar] [CrossRef] [Green Version]
- Sheng, X.; Watanabe, A.; Li, A.; Kim, E.; Song, C.; Murata, K.; Song, D.; Minagawa, J.; Liu, Z. Structural insight into light harvesting for photosystem II in green algae. Nat. Plants 2019, 5, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Su, X.; Cao, P.; Liu, X.; Chang, W.; Li, M.; Zhang, X.; Liu, Z. Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature 2016, 534, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Ma, J.; Wei, X.; Cao, P.; Zhu, D.; Chang, W.; Liu, Z.; Zhang, X.; Li, M. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 2017, 357, 815–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grinzato, A.; Albanese, P.; Marotta, R.; Swuec, P.; Saracco, G.; Bolognesi, M.; Zanotti, G.; Pagliano, C. High-Light versus Low-Light: Effects on Paired Photosystem II Supercomplex Structural Rearrangement in Pea Plants. Int. J. Mol. Sci. 2020, 21, 8643. [Google Scholar] [CrossRef]
- Van Bezouwen, L.S.; Caffarri, S.; Kale, R.; Kouřil, R.; Thunnissen, A.M.W.H.; Oostergetel, G.T.; Boekema, E.J. Subunit and chlorophyll organization of the plant photosystem II supercomplex. Nat. Plants 2017, 3, 17080. [Google Scholar] [CrossRef]
- Kyle, D.J. The 32000 Dalton Qb Protein of Photosystem II. Photochem. Photobiol. 1985, 41, 107–116. [Google Scholar] [CrossRef]
- Forouzesh, A.; Zand, E.; Soufizadeh, S.; Samadi Foroushani, S. Classification of herbicides according to chemical family for weed resistance management strategies-an update. Weed Res. 2015, 55, 334–358. [Google Scholar] [CrossRef]
- Oettmeier, W. Herbicide resistance and supersensitivity in photosystem II. Cell. Mol. Life Sci. 1999, 55, 1255–1277. [Google Scholar] [CrossRef] [PubMed]
- Trebst, A. Inhibitors in the functional dissection of the photosynthetic electron transport system. Photosynth. Res. 2007, 92, 217–224. [Google Scholar] [CrossRef]
- Broser, M.; Glöckner, C.; Gabdulkhakov, A.; Guskov, A.; Buchta, J.; Kern, J.; Müh, F.; Dau, H.; Saenger, W.; Zouni, A. Structural basis of cyanobacterial photosystem II inhibition by the herbicide terbutryn. J. Biol. Chem. 2011, 286, 15964–15972. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, C.R.D.; Bibikova, M.V.; Sabatino, P.; Oesterhelt, D.; Michel, H. Structural basis of the drastically increased initial electron transfer rate in the reaction center from a Rhodopseudomonas viridis mutant described at 2.00-Å resolution. J. Biol. Chem. 2000, 275, 39364–39368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katona, G.; Snijder, A.; Gourdon, P.; Andréasson, U.; Hansson, Ö.; Andréasson, L.E.; Neutze, R. Conformational regulation of charge recombination reactions in a photosynthetic bacterial reaction center. Nat. Struct. Mol. Biol. 2005, 12, 630–631. [Google Scholar] [CrossRef]
- Lancaster, C.R.D.; Michel, H. Refined crystal structures of reaction centres from Rhodopseudomonas viridis in complexes with the herbicide atrazine and two chiral atrazine derivatives also lead to a new model of the bound carotenoid. J. Mol. Biol. 1999, 286, 883–898. [Google Scholar] [CrossRef]
- Michel, H.; Deisenhofer, J. Relevance of the photosynthetic reaction center from purple bacteria to the structure of photosystem II. Biochemistry 1988, 27, 1–7. [Google Scholar] [CrossRef]
- Shaner, D.L. Herbicide Handbook, 10th ed.; Champaign (Ill.); Weed Science Society of America: Westminster, CO, USA, 2014; ISBN 9780615989372. [Google Scholar]
- Devine, M.D.; Shukla, A. Altered target sites as a mechanism of herbicide resistance. Crop Prot. 2000, 19, 881–889. [Google Scholar] [CrossRef]
- Powles, S.B.; Yu, Q. Evolution in Action: Plants Resistant to Herbicides. Annu. Rev. Plant Biol. 2010, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckie, H.J.; Tardif, F.J. Herbicide cross resistance in weeds. Crop Prot. 2012, 35, 15–28. [Google Scholar] [CrossRef]
- Ohad, N.; Hirschberg, J. Mutations in the D1 subunit of photosystem II distinguish between quinone and herbicide binding sites. Plant Cell 1992, 4, 273–282. [Google Scholar] [CrossRef] [Green Version]
- Draber, W.; Tietjen, K.; Kluth, J.F.; Trebst, A. Herbicides in Photosynthesis Research. Angew. Chem. Int. Ed. Engl. 1991, 30, 1621–1633. [Google Scholar] [CrossRef]
- Takahashi, R.; Hasegawa, K.; Takano, A.; Noguchi, T. Structures and binding sites of phenolic herbicides in the QB pocket of photosystem II. Biochemistry 2010, 49, 5445–5454. [Google Scholar] [CrossRef]
- Cutulle, M.A.; Armel, G.R.; Brosnan, J.T.; Best, M.D.; Kopsell, D.A.; Bruce, B.D.; Bostic, H.E.; Layton, D.S. Synthesis and evaluation of heterocyclic analogues of bromoxynil. J. Agric. Food Chem. 2014, 62, 329–336. [Google Scholar] [CrossRef]
- Pfister, K.; Arntzen, C.J. The mode of Action of Photosystem II-Specific Inhibitors in Herbicide-Resistant Weed Biotypes. Z. Naturforsch. Sect. C J. Biosci. 1979, 34, 996–1009. [Google Scholar] [CrossRef]
- Hirschberg, J.; Mcintosh, L. Molecular Basis of Herbicide Resistance in Amaranthus hybridus. Science 1983, 222, 1346–1349. [Google Scholar] [CrossRef]
- Trebst, A. The three-dimensional structure of the herbicide binding niche on the reaction center polypeptides of photosystem ii. Z. Naturforsch. Sect. C J. Biosci. 1987, 42, 742–750. [Google Scholar] [CrossRef]
- Sadekar, S.; Raymond, J.; Blankenship, R.E. Conservation of distantly related membrane proteins: Photosynthetic reaction centers share a common structural core. Mol. Biol. Evol. 2006, 23, 2001–2007. [Google Scholar] [CrossRef] [Green Version]
- Guskov, A.; Kern, J.; Gabdulkhakov, A.; Broser, M.; Zouni, A.; Saenger, W. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 2009, 16, 334–342. [Google Scholar] [CrossRef]
- Walker, D.A. ‘And whose bright presence’—An appreciation of Robert Hill and his reaction. Photosynth. Res. 2002, 73, 51–54. [Google Scholar] [CrossRef] [PubMed]
- Koblížek, M.; Malý, J.; Masojídek, J.; Komenda, J.; Kučera, T.; Giardi, M.T.; Mattoo, A.K.; Pilloton, R. A biosensor for the detection of triazine and phenylurea herbicides designed using Photosystem II coupled to a screen-printed electrode. Biotechnol. Bioeng. 2002, 78, 110–116. [Google Scholar] [CrossRef]
- Stirbet, A. Govindjee On the relation between the Kautsky effect (chlorophyll a fluorescence induction) and Photosystem II: Basics and applications of the OJIP fluorescence transient. J. Photochem. Photobiol. B Biol. 2011, 104, 236–257. [Google Scholar] [CrossRef] [PubMed]
- Hassannejad, S.; Lotfi, R.; Ghafarbi, S.P.; Oukarroum, A.; Abbasi, A.; Kalaji, H.M.; Rastogi, A. Early identification of herbicide modes of action by the use of chlorophyll fluorescence measurements. Plants 2020, 9, 529. [Google Scholar] [CrossRef]
- Buonasera, K.; Lambreva, M.; Rea, G.; Touloupakis, E.; Giardi, M.T. Technological applications of chlorophyll a fluorescence for the assessment of environmental pollutants. Anal. Bioanal. Chem. 2011, 401, 1139–1151. [Google Scholar] [CrossRef]
- Hiraki, M.; Van Rensen, J.J.S.; Vredenberg, W.J.; Wakabayashi, K. Characterization of the alterations of the chlorophyll a fluorescence induction curve after addition of Photosystem II inhibiting herbicides. Photosynth. Res. 2003, 78, 35–46. [Google Scholar] [CrossRef]
- Rea, G.; Polticelli, F.; Antonacci, A.; Scognamiglio, V.; Katiyar, P.; Kulkarni, S.A.; Johanningmeier, U.; Giardi, M.T. Structure-based design of novel Chlamydomonas reinhardtii D1-D2 photosynthetic proteins for herbicide monitoring. Protein Sci. 2009, 18, 2139–2151. [Google Scholar] [CrossRef] [Green Version]
- Christensen, M.G.; Teicher, H.B.; Streibig, J.C. Linking fluorescence induction curve and biomass in herbicide screening. Pest Manag. Sci. 2003, 59, 1303–1310. [Google Scholar] [CrossRef]
- Ventrella, A.; Catucci, L.; Agostiano, A. Herbicides affect fluorescence and electron transfer activity of spinach chloroplasts, thylakoid membranes and isolated Photosystem II. Bioelectrochemistry 2010, 79, 43–49. [Google Scholar] [CrossRef]
- Pfister, K.; Radosevich, S.R.; Arntzen, C.J. Modification of Herbicide Binding to Photosystem II in Two Biotypes of Senecio vulgaris L. Plant Physiol. 1979, 64, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, R.; Fuerst, E.P.; Nakatani, H.Y.; Arntzen, C.J. A second site for herbicide action in Photosystem II. BBA Bioenerg. 1985, 808, 293–299. [Google Scholar] [CrossRef]
- Mine, A.; Matsunaka, S. Mode of action of bentazon: Effect on photosynthesis. Pestic. Biochem. Physiol. 1975, 5, 444–450. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Zobnina, V.; Lambreva, M.D.; Rea, G.; Campi, G.; Antonacci, A.; Scognamiglio, V.; Giardi, M.T.; Polticelli, F. The plastoquinol–plastoquinone exchange mechanism in photosystem II: Insight from molecular dynamics simulations. Photosynth. Res. 2017, 131, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Reifler, M.J.; Szalai, V.A.; Peterson, C.N.; Brudvig, G.W. Effects of tail-like substituents on the binding of competitive inhibitors to the QB site of photosystem II. J. Mol. Recognit. 2001, 14, 157–165. [Google Scholar] [CrossRef]
- Krieger-Liszkay, A.; Rutherford, A.W. Influence of herbicide binding on the redox potential of the quinone acceptor in photosystem II: Relevance to photodamage and phytotoxicity. Biochemistry 1998, 37, 17339–17344. [Google Scholar] [CrossRef]
- Giardi, M.T.; Scognamiglio, V.; Rea, G.; Rodio, G.; Antonacci, A.; Lambreva, M.; Pezzotti, G.; Johanningmeier, U. Optical biosensors for environmental monitoring based on computational and biotechnological tools for engineering the photosynthetic D1 protein of Chlamydomonas reinhardtii. Biosens. Bioelectron. 2009, 25, 294–300. [Google Scholar] [CrossRef]
- Carvalho, F.P. Pesticides, environment, and food safety. Food Energy Secur. 2017, 6, 48–60. [Google Scholar] [CrossRef]
- Murphy, B.P.; Tranel, P.J. Target-Site Mutations Conferring Herbicide Resistance. Plants 2019, 8, 382. [Google Scholar] [CrossRef] [Green Version]
- Hewitt, E.J. Sand and Water Culture Methods Used in the Study of Plant Nutrition; Commonwealth Agricultural Bureaux: Farnham Royal, UK, 1966. [Google Scholar]
- Pagliano, C.; Barera, S.; Chimirri, F.; Saracco, G.; Barber, J. Comparison of the α and β isomeric forms of the detergent n-dodecyl-D-maltoside for solubilizing photosynthetic complexes from pea thylakoid membranes. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 1506–1515. [Google Scholar] [CrossRef] [Green Version]
- Porra, R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002, 73, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Pagliano, C.; Raviolo, M.; Dalla Vecchia, F.; Gabbrielli, R.; Gonnelli, C.; Rascio, N.; Barbato, R.; La Rocca, N. Evidence for PSII donor-side damage and photoinhibition induced by cadmium treatment on rice (Oryza sativa L.). J. Photochem. Photobiol. B Biol. 2006, 84, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]



| Herbicide | Chemical Class | Classification (HRAC/WSSA)/Site of Action | Molecular Structure |
|---|---|---|---|
| Diuron | Urea | 5/Ser264 |  |
| Metobromuron | Urea | 5/Ser264 |  |
| Terbuthylazine | Triazine | 5/Ser264 |  |
| Metribuzin | Triazinone | 5/Ser264 |  |
| Bentazon | Benzothiadiazinone | 6/His215 |  |
| Herbicide | DPIP Photoreduction | OJIP Fluorescence |
|---|---|---|
| I50 (M) | I50 (M) | |
| Diuron | 7.18 × 10−8 ± 1.53 × 10−9 | 8.02 × 10−8 ± 5.67 × 10−10 |
| Terbuthylazine | 1.10 × 10−7 ± 2.75 × 10−9 | 1.39 × 10−7 ± 1.60 × 10−9 |
| Bentazon | 6.16 × 10−6 ± 3.89 × 10−7 | 6.70 × 10−6 ± 8.30 × 10−8 |
| Metribuzin | 1.56 × 10−7 ± 5.95 × 10−9 | 1.24 × 10−7 ± 1.13 × 10−9 |
| Metobromuron | 1.53 × 10−6 ± 7.29 × 10−8 | 1.02 × 10−6 ± 1.03 × 10−8 |
| Herbicide | Molecular Docking | |
|---|---|---|
| ∆Gint (kcal/mol) | ∆Gdiss (kcal/mol) | |
| Diuron | −2.2 | 1.4 |
| Terbuthylazine | −1.4 | 1.5 |
| Bentazon | −0.5 | −0.1 |
| Metribuzin | −1.5 | 1.1 |
| Metobromuron | −1.1 | 0.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Battaglino, B.; Grinzato, A.; Pagliano, C. Binding Properties of Photosynthetic Herbicides with the QB Site of the D1 Protein in Plant Photosystem II: A Combined Functional and Molecular Docking Study. Plants 2021, 10, 1501. https://0-doi-org.brum.beds.ac.uk/10.3390/plants10081501
Battaglino B, Grinzato A, Pagliano C. Binding Properties of Photosynthetic Herbicides with the QB Site of the D1 Protein in Plant Photosystem II: A Combined Functional and Molecular Docking Study. Plants. 2021; 10(8):1501. https://0-doi-org.brum.beds.ac.uk/10.3390/plants10081501
Chicago/Turabian StyleBattaglino, Beatrice, Alessandro Grinzato, and Cristina Pagliano. 2021. "Binding Properties of Photosynthetic Herbicides with the QB Site of the D1 Protein in Plant Photosystem II: A Combined Functional and Molecular Docking Study" Plants 10, no. 8: 1501. https://0-doi-org.brum.beds.ac.uk/10.3390/plants10081501