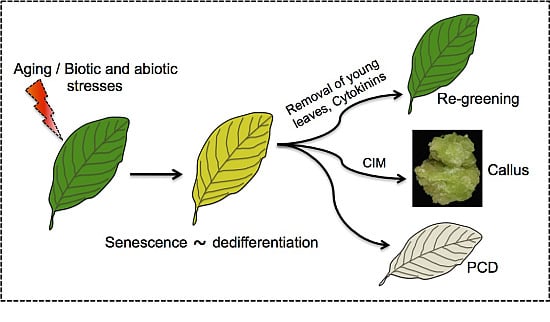
Senescence Meets Dedifferentiation
Abstract
:1. Introduction
2. Reversal of Leaf Senescence
3. What Is Dedifferentiation?
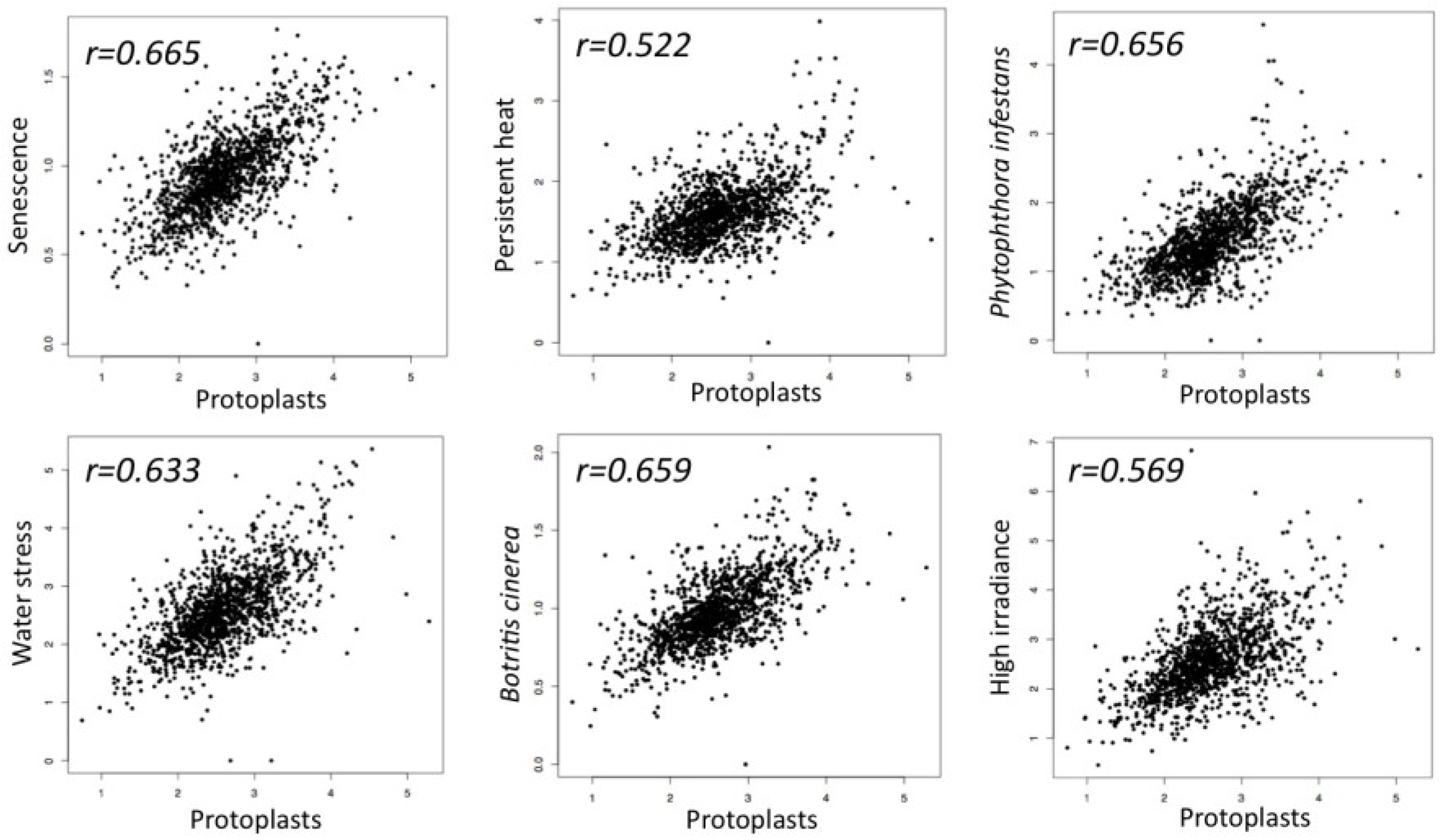
4. Senescence Meets Dedifferentiation
4.1. Chromatin Structure
4.2. Ribosome Biogenesis and Protein Synthesis
4.3. Activation of Transposable Elements (TEs)
5. Concluding Remarks

Acknowledgements
Author Contributions
Conflicts of Interest
References
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf senescence—A genomics approach. Plant Biotechnol. J. 2003, 1, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Gepstein, S. Leaf senescence—Not just a “wear and tear” phenomenon. Genome Biol. 2004, 5, 212. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.F.; Wu, S.H. Molecular events in senescing Arabidopsis leaves. Plant J. 2004, 39, 612–628. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cai, Z.; Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004, 27, 521–549. [Google Scholar] [CrossRef]
- Andersson, A.; Keskitalo, J.; Sjödin, A.; Bhalerao, R.; Sterky, F.; Wissel, K.; Tandre, K.; Aspeborg, H.; Moyle, R.; Ohmiya, Y.; et al. A transcriptional timetable of autumn senescence. Genome Biol. 2004, 5, R24. [Google Scholar] [CrossRef] [PubMed]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.S.; Penfold, C.A.; Jenkins, D.; et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S.; Holopainen, J.K. Why red-dominated autumn leaves in America and yellow-dominated autumn leaves in Northern Europe? New Phytol. 2009, 183, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Matile, P. Biochemistry of Indian summer: Physiology of autumnal leaf coloration. Exp. Gerontol. 2000, 35, 145–158. [Google Scholar] [CrossRef]
- Hoch, W.A.; Zeldin, E.L.; McCown, B.H. Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiol. 2001, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Archetti, M.; Döring, T.F.; Hagen, S.B.; Hughes, N.M.; Leather, S.R.; Lee, D.W.; Lev-Yadun, S.; Manetas, Y.; Ougham, H.J.; Schaberg, P.G.; et al. Unravelling the evolution of autumn colours: An interdisciplinary approach. Trends Ecol. Evol. 2009, 24, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, K.A.; Bennett, A.B. Programmed senescence of plant organs. Cell Death Differ. 1997, 4, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Jibran, R.; Hunter, D.A.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Zacarias, L.; Reid, M.S. Role of growth regulators in the senescence of Arabidopsis thaliana leaves. Physiol. Plant. 1990, 80, 549–554. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112. [Google Scholar] [PubMed]
- Kim, J.; Chang, C.; Tucker, M.L. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Richmond, A.E.; Lang, A. Effect of kinetin on protein content and survival of detached Xanthium leaves. Science 1957, 125, 650–651. [Google Scholar] [CrossRef]
- Goldthwaite, J.J.; Laetsch, W.M. Control of senescence in Rumex leaf discs by gibberellic acid. Plant Physiol. 1968, 43, 1855–1858. [Google Scholar] [CrossRef] [PubMed]
- Nooden, L.D.; Kahanak, G.M.; Okatan, Y. Prevention of monocarpic senescence in soybeans with auxin and cytokinin: An antidote for self-destruction. Science 1979, 206, 841–843. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef] [PubMed]
- Ori, N.; Juarez, M.T.; Jackson, D.; Yamaguchi, J.; Banowetz, G.M.; Hake, S. Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence—Activated promoter. Plant Cell 1999, 11, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Mandahar, C.L.; Suri, R.A. Cytokinin-like activity in green islands formed around infection spots of Alternaria brassicicola on mustard leaves in nature and their role in pathogenesis. Biol. Plant. 1987, 29, 76–79. [Google Scholar] [CrossRef]
- Giron, D.; Kaiser, W.; Imbault, N.; Casas, J. Cytokinin-mediated leaf manipulation by a leafminer caterpillar. Biol. Lett. 2007, 3, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.A.; Osborne, D.J. Gibberellin, as a regulator of protein and ribonucleic acid synthesis during senescence in leaf cells of Taraxacum officinale. Can. J. Bot. 1966, 44, 739–745. [Google Scholar] [CrossRef]
- Beevers, L. Effect of gibberellic acid on the senescence of leaf discs of Nasturtium (Tropaeolum majus). Plant Physiol. 1966, 41, 1074–1076. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 2013, 197, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Wittenbach, V.A. Induced senescence of intact wheat seedlings and its reversibility. Plant Phys. 1977, 59, 1038–1042. [Google Scholar]
- Greening, M.T.; Butterfield, F.J.; Harris, N. Chloroplast ultrastructure during senescence and regreening of flax cotyledons. New Phytol. 1982, 92, 279–285. [Google Scholar] [CrossRef]
- Zavaleta-Mancera, H.A.; Thomas, B.J.; Thomas, H.; Scott, I.M. Regreening of senescent Nicotiana leaves. II. Redifferentiation of plastids. J. Exp. Bot. 1999, 50, 1683–1689. [Google Scholar]
- Girardin, P.; Deltour, A.; Tollenaar, M. Effect of temporary N starvation in maize on leaf senescence. Can. J. Plant Sci. 1985, 65, 819–829. [Google Scholar] [CrossRef]
- Schildhauer, J.; Wiedemuth, K.; Humbeck, K. Supply of nitrogen can reverse senescence processes and affect expression of genes coding for plastidic glutamine synthetase and lysine-ketoglutarate reductase/saccharopine dehydrogenase. Plant Biol. 2008, 1, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Schildhauer, J.; Araújo, W.L.; Munné-Bosch, S.; Fernie, A.R.; Proost, S.; Humbeck, K.; Mueller-Roeber, B. Reversal of senescence by N resupply to N-starved Arabidopsis thaliana: Transcriptomic and metabolomic consequences. J. Exp. Bot. 2014, 65, 3975–3992. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.; Ougham, H.J.; Wagstaff, C.; Stead, A.D. Defining senescence and death. J. Exp. Bot. 2003, 54, 1127–1132. [Google Scholar] [CrossRef] [PubMed]
- Theise, N.D. New principles of cell plasticity. C. R. Biol. 2002, 325, 1039–1043. [Google Scholar] [CrossRef]
- Bloom, W. Cellular differentiation and tissue culture. Physiol. Rev. 1937, 17, 589–617. [Google Scholar]
- Thornton, C.S. The histogenesis of muscle in the regenerating fore limb of larval Amblystoma punctatum. J. Morph. 1938, 62, 17–47. [Google Scholar] [CrossRef]
- Hay, E.D. Electron microscopic observations of muscle dedifferentiation in regenerating Amblystoma limbs. Dev. Biol. 1959, 1, 555–585. [Google Scholar] [CrossRef]
- Grafi, G.; Avivi, Y. Stem cells: A lesson from dedifferentiation. Trends Biotechnol. 2004, 22, 388–389. [Google Scholar] [CrossRef] [PubMed]
- McKay, R. Stem cells—Hype and hope. Nature 2000, 406, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Morozova, N.; Williams, L.; Libs, L.; Avivi, Y.; Grafi, G. Two phases of chromatin decondensation during dedifferentiation of plant cells: Distinction between competence for cell fate switch and a commitment for S phase. J. Biol. Chem. 2001, 276, 22772–22778. [Google Scholar] [CrossRef] [PubMed]
- Grafi, G. How cells dedifferentiate: A lesson from plants. Dev. Biol. 2004, 268, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Damri, M.; Granot, G.; Ben-Meir, H.; Avivi, Y.; Plaschkes, I.; Chalifa-Caspi, V.; Wolfson, M.; Fraifeld, V.; Grafi, G. Senescing cells share common features with dedifferentiating cells. Rejuvenation Res. 2009, 12, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Grafi, G.; Chalifa-Caspi, V.; Nagar, T.; Plaschkes, I.; Barak, S.; Ransbotyn, V. Plant response to stress meets dedifferentiation. Planta 2011, 233, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Gaspar-Maia, A.; Alajem, A.; Meshorer, E.; Ramalho-Santos, M. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011, 12, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Grafi, G.; Florentin, A.; Ransbotyn, V.; Morgenstern, Y. The stem cell state in plant development and in response to stress. Front. Plant Sci. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- MacRae, E.K.; Meetz, G.D. Electron microscopy of the ammoniacal silver reaction for histones in the erythropoietic cells of the chick. J. Cell Biol. 1970, 45, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.J., Jr.; Bertles, J.F.; Gordon, A.S. Identifying characteristics of the haematopoietic precursor cell. J. Cell Sci. 1971, 9, 23–47. [Google Scholar] [PubMed]
- Miura, A.B.; Shibata, A.; Akihama, T.; Endo, Y.; Saito, Y. Ultrastructure of developing erythrocytes. Tohoku J. Exp. Med. 1974, 112, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978, 5, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.J.; Rhodes, D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 2006, 16, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Booker, C.E.; Dwivedi, R.S. Ultrastructure of meristematic cells of dormant and released buds in Tradescantia paludosa. Exp. Cell Res. 1973, 82, 255–261. [Google Scholar] [CrossRef]
- Yadav, R.K.; Girke, T.; Pasala, S.; Xie, M.; Reddy, G.V. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 2009, 106, 4941–4946. [Google Scholar] [CrossRef] [PubMed]
- Ay, N.; Irmler, K.; Fischer, A.; Uhlemann, R.; Reuter, G.; Humbeck, K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 2009, 58, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Florentin, A.; Damri, M.; Grafi, G. Stress induces plant somatic cells to acquire some features of stem cells accompanied by selective chromatin reorganization. Dev. Dyn. 2013, 242, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Pecinka, A.; Dinh, H.Q.; Baubec, T.; Rosa, M.; Lettner, N.; Mittelsten Scheid, O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 2010, 22, 3118–3129. [Google Scholar] [CrossRef] [PubMed]
- Grafi, G.; Barak, S. Stress induces cell dedifferentiation in plants. Biochim. Biophys. Acta 2015, 1849, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Halicka, H.D.; Zhao, H.; Podhorecka, M.; Traganos, F.; Darzynkiewicz, Z. Cytometric detection of chromatin relaxation, an early reporter of DNA damage response. Cell Cycle 2009, 8, 2233–2237. [Google Scholar] [CrossRef] [PubMed]
- Abrahan, C.E.; Insua, M.F.; Politi, L.E.; German, O.L.; Rotstein, N.P. Oxidative stress promotes proliferation and dedifferentiation of retina glial cells in vitro. J. Neurosci. Res. 2009, 87, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Shoshani, O.; Zipori, D. Mammalian cell dedifferentiation as a possible outcome of stress. Stem Cell Rev. 2011, 7, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Shoshani, O.; Zipori, D. Stress as a fundamental theme in cell plasticity. Biochim. Biophys. Acta 2015, 1849, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.; Zhao, J.; Morozova, N.; Li, Y.; Avivi, Y.; Grafi, G. Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev. Dyn. 2003, 228, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Krul, W.R. Nucleic acid and protein metabolism of senescing and regenerating soybean cotyledons. Plant Physiol. 1974, 54, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Makrides, S.C.; Goldthwaite, J. Biochemical changes during bean leaf growth, maturity and senescence: Contents of DNA, polyribosomes, ribosomal RNA, protein and chlorophyll. J. Exp. Bot. 1981, 32, 725–735. [Google Scholar] [CrossRef]
- Skadsen, R.W.; Cherry, J.H. Quantitative changes in in vitro and in vivo protein synthesis in aging and rejuvenated soybean cotyledons. Plant Physiol. 1983, 71, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, B.I.; Arglebe, C. Studies on ribosomes from barley leaves: Changes during senescence. Plant Physiol. 1967, 42, 1497–503. [Google Scholar] [CrossRef] [PubMed]
- Eilam, Y.; Butler, R.D.; Simon, E.W. Ribosomes and polysomes in cucumber leaves during growth and senescence. Plant Physiol. 1971, 47, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Steeves, T.A.; Sussex, I.M. Analytical studies of the shoot apex. In Patterns in Plant Development; Cambridge University Press: Cambridge, NY, USA, 1989; pp. 62–85. [Google Scholar]
- Cheung, T.H.; Rando, T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013, 14, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Kidwell, M.G.; Lisch, D.R. Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution 2001, 55, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. Epigenetic regulation of transposable elements in plants. Annu. Rev. Plant Biol. 2009, 60, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Grandbastien, M.A. LTR retrotransposons, handy hitchhikers of plant regulation and stress response. Biochim. Biophys. Acta 2015, 1849, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Kuo, D.; Nathanson, J.; Satoh, A.; Pao, G.M.; Yeo, G.W.; Bryant, S.V.; Voss, S.R.; Gardiner, D.M.; Hunter, T. Retrotransposon long interspersed nucleotide element-1 (LINE-1) is activated during salamander limb regeneration. Dev. Growth Differ. 2012, 54, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.R.; Kumar, A.; Flavell, A.J. Activation of the Ty1-copia group retrotransposons of potato (Solanum tuberosum) during protoplast isolation. Plant Cell Rep. 1996, 15, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Pouteau, S.; Huttner, E.; Grandbastien, M.A.; Caboche, M. Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 1991, 10, 1911–1918. [Google Scholar] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Melayah, D.; Bonnivard, E.; Chalhoub, B.; Audeon, C.; Grandbastien, M.A. The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J. 2001, 28, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Gepstein, S.; Glick, B.R. Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 2013, 82, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.; Pearce, S.P.; Distelfeld, A.; Christiansen, M.W.; Uauy, C.; Akhunov, E.; Fahima, T.; Dubcovsky, J. Effect of the down-regulation of the high Grain Protein Content (GPC) genes on the wheat transcriptome during monocarpic senescence. BMC Genomics 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Eaton, C.J.; Cox, M.P.; Ambrose, B.; Becker, M.; Hesse, U.; Schardl, C.L.; Scott, B. Disruption of signaling in a fungal-grass symbiosis leads to pathogenesis. Plant Physiol. 2010, 153, 1780–1794. [Google Scholar] [CrossRef] [PubMed]
- Bundock, P.; Hooykaas, P. An Arabidopsis hAT-like transposase is essential for plant development. Nature 2005, 436, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Macia, A.; Blanco-Jimenez, E.; García-Pérez, J.L. Retrotransposons in pluripotent cells: Impact and new roles in cellular plasticity. Biochim. Biophys. Acta 2015, 1849, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Perez, J.L.; Marchetto, M.C.; Muotri, A.R.; Coufal, N.G.; Gage, F.H.; O’Shea, K.S.; Moran, J.V. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007, 16, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Wissing, S.; Muñoz-Lopez, M.; Macia, A.; Yang, Z.; Montano, M.; Collins, W.; Garcia-Perez, J.L.; Moran, J.V.; Greene, W.C. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum. Mol. Genet. 2012, 21, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Wallace, N.A.; Belancio, V.P.; Deininger, P. L1 mobile element expression causes multiple types of toxicity. Gene 2008, 419, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Belgnaoui, S.M.; Gosden, R.G.; Semmes, O.J.; Haoudi, A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006, 6. [Google Scholar] [CrossRef] [PubMed]
- Gasior, S.L.; Wakeman, T.P.; Xu, B.; Deininger, P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006, 357, 1383–1393. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Criscione, S.W.; Peckham, E.J.; Hillenmeyer, S.; Hamm, E.A.; Manivannan, J.; Peterson, A.L.; Kreiling, J.A.; Neretti, N.; Sedivy, J.M. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 2013, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Geesman, G.J.; Hostikka, S.L.; Atallah, M.; Blackwell, B.; Lee, E.; Cook, P.J.; Pasaniuc, B.; Shariat, G.; Halperin, E.; et al. Inhibition of activated pericentromeric SINE/Alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 2011, 10, 3016–3030. [Google Scholar] [CrossRef] [PubMed]
- Tessadori, F.; Chupeau, M.C.; Chupeau, Y.; Knip, M.; Germann, S.; van Driel, R.; Fransz, P.; Gaudin, V. Large-scale dissociation and sequential reassembly of pericentric heterochromatin in dedifferentiated Arabidopsis cells. J. Cell Sci. 2007, 120, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Grafi, G. Stress cycles in stem cells/iPSCs development: Implications for tissue repair. Biogerontology 2013, 14, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Larkin, P.J.; Scowcroft, W.R. Somaclonal variation—A novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rapp, Y.G.; Ransbotyn, V.; Grafi, G. Senescence Meets Dedifferentiation. Plants 2015, 4, 356-368. https://0-doi-org.brum.beds.ac.uk/10.3390/plants4030356
Rapp YG, Ransbotyn V, Grafi G. Senescence Meets Dedifferentiation. Plants. 2015; 4(3):356-368. https://0-doi-org.brum.beds.ac.uk/10.3390/plants4030356
Chicago/Turabian StyleRapp, Yemima Givaty, Vanessa Ransbotyn, and Gideon Grafi. 2015. "Senescence Meets Dedifferentiation" Plants 4, no. 3: 356-368. https://0-doi-org.brum.beds.ac.uk/10.3390/plants4030356