Keeping Control: The Role of Senescence and Development in Plant Pathogenesis and Defense
Abstract
:1. Introduction
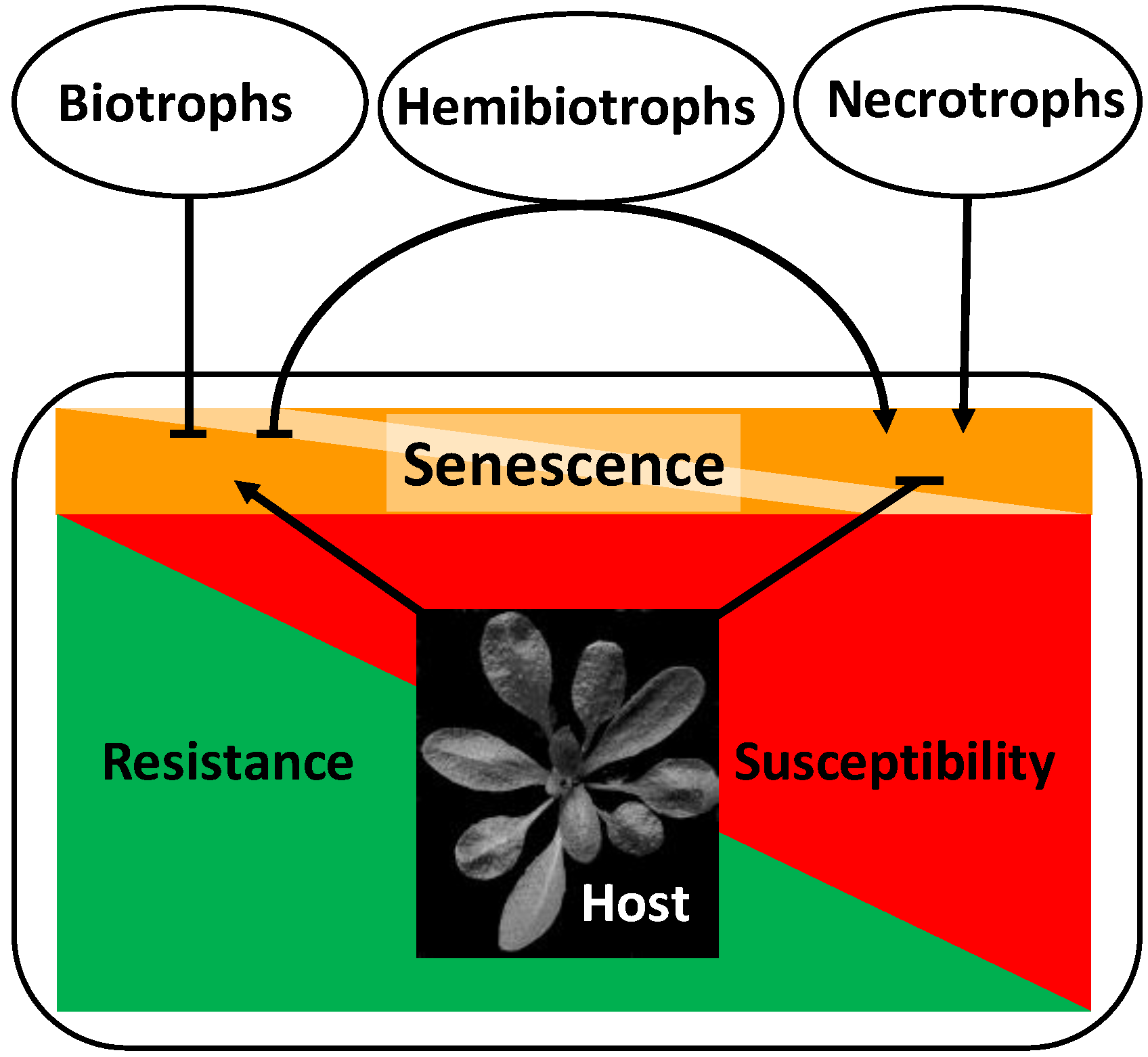
2. Major Players in the Control of Senescence and Pathogen Response
2.1. Phytohormones
2.1.1. Ethylene
2.1.2. Jasmonic Acid
2.1.3. Salicylic Acid
2.1.4. Abscisic Acid
2.1.5. Brassinosteroids
2.1.6. Cytokinins, Auxins, Gibberellins
2.2. Transcription Factors
2.3. Reactive Oxygen Species, Programmed Cell Death and Autophagy
3. Developmental Implications of Host-Pathogen-Interactions
3.1. Host Development Affects Pathogenesis
3.1.1. Senescence-Like Processes Confer Quantitative Resistance against Biotrophic Pathogens in Cereals
3.1.2. Late-Senescing Genotypes Are More Resistant to Necrotrophic and Hemibiotrophic Pathogens

3.2. Pathogenesis Affects Host Development
3.2.1. Pathogens Manipulate Host Development for Their Own Benefit
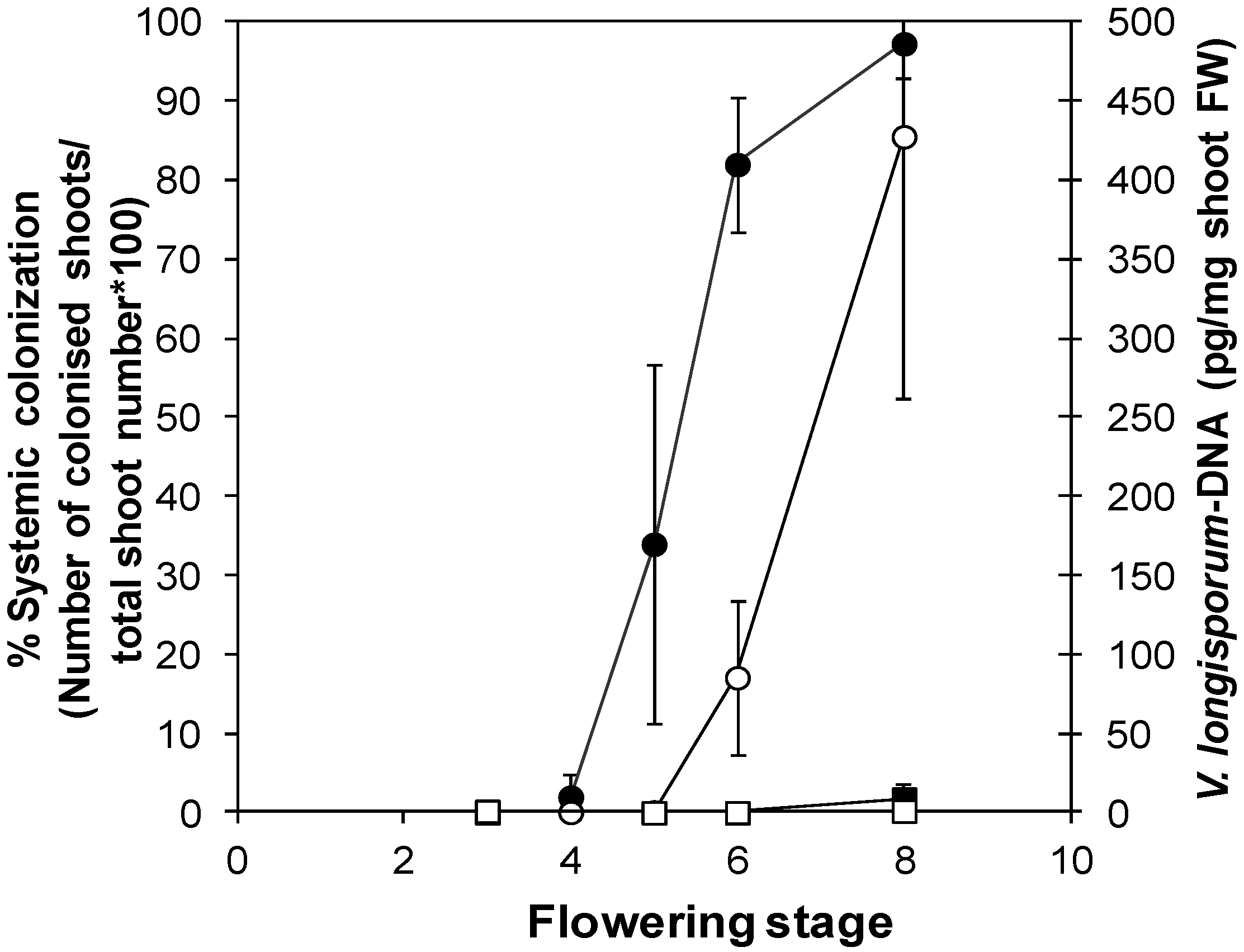
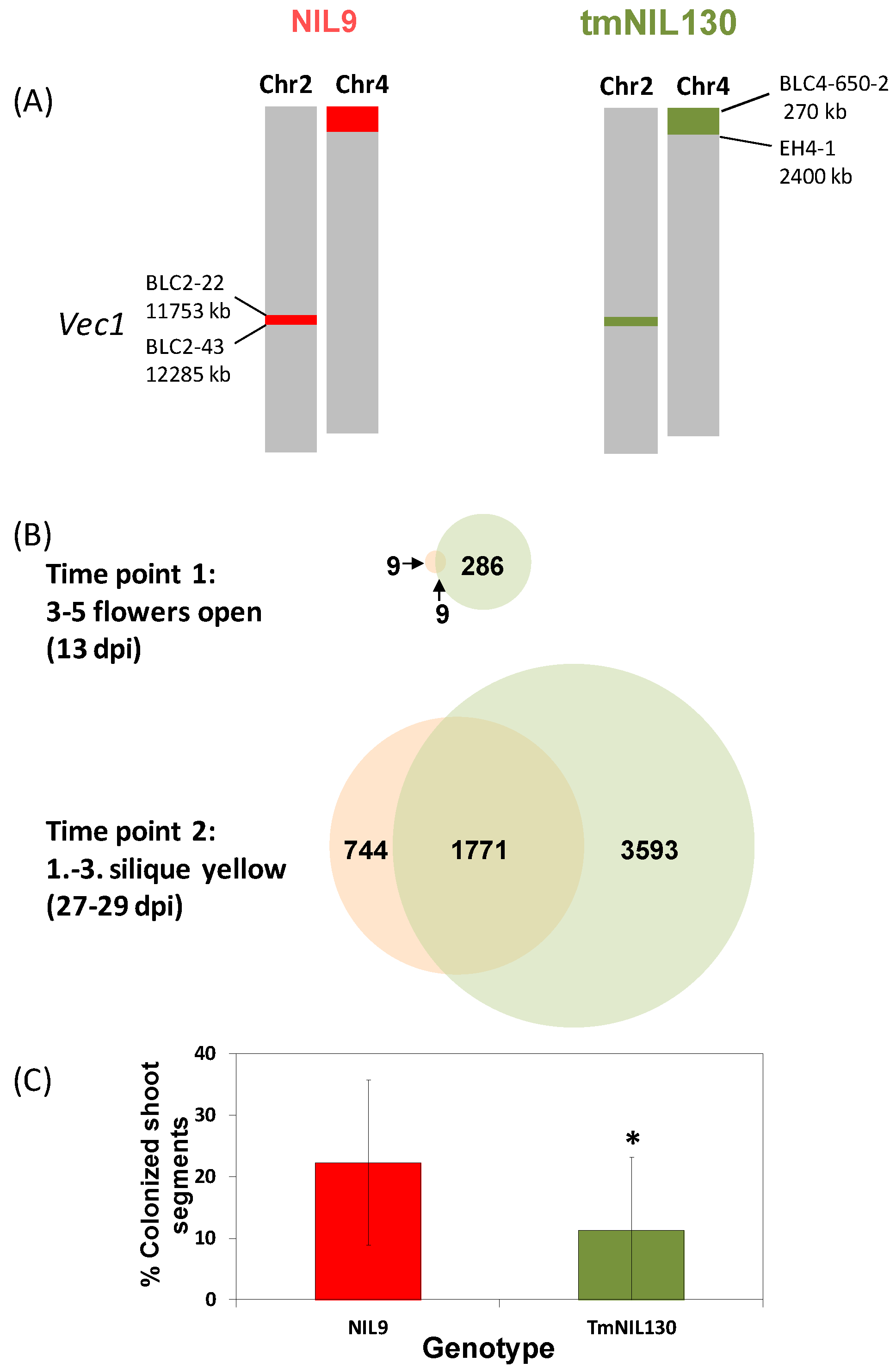
3.2.2. “Developmental Buffering” Confers Quantitative Resistance to V. longisporum in A. thaliana

3.2.3. Transcriptional Response to V. longisporum at Maturity in a Susceptible and a Resistant A. thaliana Line

| NIL9 | tmNIL130 | ||||
|---|---|---|---|---|---|
| Gene | FC | Gene | FC | Gene | FC |
| Ethylene biosynthesis, -signaling and -response | |||||
| at2g25450 | 6.94 | at5g20400 | 2.13 | at5g44210 (ERF9) | 2.59 |
| at4g37770 (ACS8) | 4.07 | at5g59540 | 2.48 | at3g23150 (ETR2) | 2.46 |
| at5g65800 (ACS5) | 3.90 | at5g43440 | 2.18 | at1g04310 (ERS2) | 2.84 |
| at1g01480(ACS2) | 3.59 | at1g03400 | 2.10 | at3g50260 (CEJ1) | 2.84 |
| at4g26200 (ACS7) | 3.18 | at5g43450 | 2.13 | at2g40940 (ERS1) | 2.40 |
| at5g44210 (ERF9) | 2.94 | at2g25450 | 12.06 | at5g25190 | 3.67 |
| at5g25190 | 5.09 | at4g37770 (ACS8) | 4.99 | at3g11930 | 2.25 |
| at4g37580 (HLS1) | 2.07 | at5g65800(ACS5) | 3.88 | at1g04370 (ATERF14) | 4.98 |
| at3g11930 | 3.67 | at1g01480 (ACS2) | 11.90 | at1g55150 | 3.49 |
| at1g55150 | 2.10 | at3g61510 (ACS1) | 5.00 | at1g09740 | 2.32 |
| at3g20640 | 2.40 | at4g26200 (ACS7) | 2.17 | ||
| at1g77330 | 0.28 | at2g31230 (ATERF15) | 0.40 | ||
| at5g07580 | 0.38 | at2g20100 | 0.45 | ||
| at2g31230 (ATERF15) | 0.42 | at1g27660 | 0.32 | ||
| at1g06160 (ORA59) | 0.33 | at1g49830 | 0.33 | ||
| at5g44350 | 0.21 | at4g29100 | 0.48 | ||
| at1g27660 | 0.37 | ||||
| Jasmonic acid biosynthesis, signaling and response | |||||
| at1g18020 | 2.89 | ||||
| at1g52070 | 2.64 | ||||
| at5g48180 (NSP5) | 2.25 | ||||
| at1g55020 (LOX1) | 0.44 | at1g09400 | 0.35 | ||
| at1g76690 (OPR2) | 0.39 | at3g16470 (JR1) | 0.24 | ||
| at3g16470 (JR1) | 0.34 | at3g16450 | 0.40 | ||
| at3g16450 | 0.35 | ||||
| Salicylic acid biosynthesis, signaling and response | |||||
| at1g66720 | 6.23 | at1g66720 | 12.03 | ||
| at5g04370 (NAMT1) | 0.18 | at5g38020 | 0.20 | at4g26420 (GAMT1) | 0.35 |
| at5g37990 | 0.09 | at1g05670 | 0.38 | at5g37990 | 0.08 |
| at5g04370 (NAMT1) | 0.45 | ||||
| Abscisic acid biosynthesis, signaling and response | |||||
| at4g18350 (NCED2) | 4.26 | at4g18350 (NCED2) | 10.1 | at5g23350 | 2.39 |
| at5g50720 (ATHVA22E) | 2.03 | at1g30100 (NCED5) | 13.32 | at5g50720 (ATHVA22E) | 2.31 |
| at1g74520 (ATHVA22A) | 2.84 | at3g24650 (ABI3) | 8.23 | ||
| at4g32810 (CCD8) | 0.41 | at2g36020 (HVA22J) | 0.20 | at2g17770 (ATBZIP27) | 0.28 |
| at2g44990 (CCD7) | 0.15 | at4g32810 (CCD8) | 0.28 | at5g62490 (ATHVA22B) | 0.49 |
| at3g63210 (MARD1) | 0.50 | at2g44990 (CCD7) | 0.16 | at1g45249 (ABF2) | 0.39 |
| at2g17770 (ATBZIP27) | 0.32 | at3g43600 (AAO2) | 0.26 | ||
| at5g23370 | 0.46 | at1g52920 (GCR2) | 0.48 | ||
| at1g45249 (ABF2) | 0.36 | at3g63210 (MARD1) | 0.45 | ||
| Brassinosteroid synthesis, signaling and response | |||||
| at3g50660 (DWF4) | 4.09 | at3g50660 (DWF4) | 2.99 | at2g22830 (SQE2) | 2.28 |
| at1g76090 (SMT3) | 2.30 | at4g36780 | 2.16 | ||
| at4g37760 (SQE3) | 0.34 | ||||
| Cytokinin biosynthesis, signaling and response | |||||
| at5g05860 (UGT76C2) | 2.34 | at5g05860 (UGT76C2) | 2.06 | at1g22400 (UGT85A1) | 2.72 |
| at2g17820 (AHK1) | 2.10 | at4g29740 (CKX4) | 5.18 | ||
| at2g47430 (CKI1) | 0.34 | at5g35750 (AHK2) | 0.45 | at2g01830 (AHK4) | 0.44 |
| at2g01830 (AHK4) | 0.44 | at2g25180 (ARR12) | 0.32 | ||
| Gibberellic acid biosynthesis, signaling and response | |||||
| at5g59845 | 6.82 | at1g02400 (ATGA2OX4) | 3.05 | ||
| at1g75750 (GASA1) | 10.66 | ||||
| at5g59845 | 5.97 | ||||
| at1g52820 | 0.24 | at4g23340 | 0.33 | at5g51810 (GA20OX2) | 0.22 |
| at4g25420 GA5 | 0.22 | at4g25420 (GA5) | 0.33 | at1g14920 (GAI) | 0.34 |
| at1g14920 (GAI) | 0.42 | ||||
| Auxin biosynthesis, signaling and response | |||||
| at5g13360 | 2.46 | at3g44300 (NIT2) | 16.35 | at5g54510 (GH3.6) | 3.00 |
| at1g10810 | 4.09 | at5g55250 (IAMT1) | 2.62 | at4g36110 | 4.11 |
| at5g13320 (PBS3) | 4.09 | at3g15450 | 6.61 | at1g17345 | 2.57 |
| at5g54510 GH3.6 | 2.78 | at2g23170 (GH3.3) | 15.67 | at4g37390 (YDK1/GH3.2) | 5.77 |
| at1g17345 | 2.48 | at1g10810 | 3.26 | at2g04850 | 2.06 |
| at2g04850 | 2.69 | at5g13320 (PBS3) | 6.08 | at4g12410 | 2.09 |
| at4g12410 | 3.37 | at1g60730 | 2.48 | at5g13370 | 4.57 |
| at5g01100 | 2.93 | at4g27450 | 3.94 | ||
| at1g77690 (LAX3) | 0.37 | at3g10870 (MES17) | 0.49 | at3g12955 | 0.34 |
| at5g47530 | 0.29 | at1g05560 (UGT1) | 0.45 | at3g02250 | 0.45 |
| at4g17280 | 0.17 | at1g77690 (LAX3) | 0.42 | at4g00880 | 0.30 |
| at1g16510 | 0.48 | at5g16530 (PIN5) | 0.42 | at1g19830 | 0.26 |
| at2g14960 | 0.24 | at1g73590 (PIN1) | 0.47 | at3g61900 | 0.45 |
| at3g02250 | 0.46 | at5g47530 | 0.21 | at1g14970 | 0.31 |
| at4g00880 | 0.46 | at4g17280 | 0.21 | at3g61750 | 0.25 |
| at3g61750 | 0.34 | at3g43120 | 0.34 | at5g65470 | 0.41 |
| at1g28130 (GH3.17) | 0.27 | at3g47620 (AtTCP14) | 0.40 | at4g34750 (SAUR_E) | 0.31 |
| at3g25290 | 0.31 | at2g04852 | 0.49 | ||
| at2g34680 (AIR9) | 0.42 | at4g12980 | 0.39 | ||
| WRKY transcription factors | |||||
| at4g31800 (WRKY18) | 2.38 | at1g69810 (WRKY36) | 2.05 | at1g69310 (WRKY57) | 1.69 |
| at5g46350 (WRKY8) | 3.85 | at4g31800 (WRKY18) | 2.76 | at3g56400 (WRKY70) | 2.48 |
| at2g25000 (WRKY60) | 4.09 | at5g46350 (WRKY8) | 4.19 | at5g64810 (WRKY51) | 4.65 |
| at1g80590 (WRKY66) | 4.13 | at1g66600 (WRKY63) | 6.33 | at1g80590 (WRKY66) | 3.18 |
| at2g47260 (WRKY23) | 4.09 | at4g23810 (WRKY53) | 3.01 | at5g26170 (ATWRKY50) | 2.96 |
| at4g22070 (WRKY31) | 3.19 | at2g23320 (WRKY15) | 2.31 | at5g49520 (WRKY48) | 1.90 |
| at4g23550 (WRKY29) | 2.83 | at2g47260 (WRKY23) | 6.37 | ||
| at2g25000 (WRKY60) | 3.91 | at4g22070 (WRKY31) | 4.67 | ||
| at1g18860 (WRKY61) | 4.38 | at1g62300 (WRKY6) | 3.73 | ||
| at5g13080 (WRKY75) | 3.47 | at5g24110 (WRKY30) | 3.15 | ||
| at5g01900 (WRKY62) | 5.49 | at1g80840 (WRKY40) | 6.85 | ||
| at5g52830 (WRKY27) | 0.45 | at4g39410 (WRKY13) | 0.50 | at2g37260 (WRKY44) | 0.34 |
| at3g58720 | 0.48 | at5g52830 (WRKY27) | 0.34 | at2g44745 | 0.47 |
| at2g34830 (WRKY35) | 0.46 | at2g34830 (WRKY35) | 0.44 | ||
| NIL9 | tmNIL130 | ||||
|---|---|---|---|---|---|
| Gene | FC | Gene | FC | Gene | FC |
| Redox homeostasis | |||||
| at1g69880 (ATH8) | 5.95 | at1g69880 (ATH8) | 7.52 | at3g02870 (VTC4) | 2.60 |
| at5g61440 (ACHT5) | 2.79 | at1g19730 (ATH4) | 3.45 | at1g63460 | 2.04 |
| at4g39830 | 4.77 | at5g61440 (ACHT5) | 3.45 | at2g31570 (ATGPX2) | 2.45 |
| at1g20630 | 2.82 | at5g16400 (TRXF2) | 2.46 | at2g16060 (GLB1) | 3.65 |
| at1g21750 (ATPDIL1-1) | 2.58 | at1g03850 | 5.63 | ||
| at3g19010 | 2.40 | at4g15700 | 4.01 | ||
| at2g43350 (ATGPX3) | 2.24 | at1g32760 | 2.01 | ||
| at4g39830 | 5.01 | at1g20630 (CAT1) | 2.62 | ||
| at4g09010 (APX4) | 3.47 | at5g17770 (CBR1) | 2.30 | ||
| at4g08390 (SAPX) | 2.54 | ||||
| at4g33040 | 0.44 | at3g20560 (ATPDIL5-3) | 0.48 | at4g33040 | 0.31 |
| at5g11930 | 0.33 | at4g18260 | 0.16 | at5g11930 | 0.26 |
| at3g19000 | 0.44 | at2g47870 | 0.41 | ||
| at5g21100 | 0.37 | at4g25100 (FSD1) | 0.23 | ||
| at2g47880 | 0.24 | at1g31170 (SRX) | 0.45 | ||
| Peroxidases | |||||
| at3g49120 (ATPERX34) | 2.51 | at5g19880 | 2.53 | at1g49570 | 5.56 |
| at2g37130 (PER21) | 4.44 | at5g19890 | 4.38 | at2g34060 | 2.20 |
| at1g71695 (PER12) | 2.29 | at3g49120 (ATPERX34) | 2.36 | at5g64110 | 4.40 |
| at5g15180 | 6.40 | at2g37130 (PER21) | 3.78 | at5g64100 | 6.12 |
| at5g64110 | 3.77 | at1g71695 (PER12) | 3.00 | at5g05340 | 3.91 |
| at5g51890 | 24.95 | at4g37520 (PER50) | 2.98 | at4g33420 | 3.14 |
| at5g64120 | 11.16 | at5g51890 | 18.62 | ||
| at5g39580 | 6.27 | at1g05240 | 2.48 | ||
| at5g15180 | 4.10 | ||||
| at5g67400 | 0.09 | at1g68850 | 0.26 | ||
| at5g14130 | 0.31 | at5g42180 (PER64) | 0.21 | ||
| at5g06720 | 0.46 | ||||
| at1g30870 | 0.33 | ||||
| at5g42180 (PER64) | 0.26 | ||||
| at1g44970 | 0.11 | ||||
| at1g05250 | 0.15 | ||||
| at1g05240 | 0.13 | ||||
| Glutathione-S-transferases | |||||
| at5g02780 | 7.93 | at1g02920 (ATGSTF7) | 4.38 | at1g14550 | 5.27 |
| at2g29440 (ATGSTU6) | 6.41 | at2g29470 (ATGSTU3) | 8.22 | at1g14540 | 6.54 |
| at3g09270: (ATGSTU8) | 3.21 | at4g19880 | 2.23 | at3g09270 (ATGSTU8) | 3.94 |
| at1g17180 (ATGSTU25) | 3.02 | at1g17170 (ATGSTU24) | 2.26 | at1g17180 (ATGSTU25) | 2.19 |
| at1g69930 (ATGSTU11) | 2.05 | at5g02780 | 3.33 | at1g69930: (ATGSTU11) | 6.67 |
| at2g29440 (ATGSTU6) | 8.74 | at1g02930 (ATGSTF6) | 5.20 | ||
| at4g02520 (ATGSTF2) | 2.74 | at2g29480 (ATGSTU2) | 4.03 | ||
| at2g02930 (ATGSTF3) | 4.33 | at2g29450 (ATGSTU5) | 2.83 | ||
| at1g69920 (ATGSTU12) | 5.13 | ||||
| at1g74590 (ATGSTU10) | 4.16 | ||||
| at1g49860 (ATGSTF14) | 0.19 | at3g03190 (ATGSTF11) | 0.27 | ||
| at1g78370 (ATGSTU20) | 0.20 | at1g78370 (ATGSTU20) | 0.24 | ||
| at1g10360 (ATGSTU18) | 0.44 | ||||
| at5g62480: (ATGSTU9) | 0.39 | ||||
| at5g17220: (ATGSTF12) | 0.10 | ||||
4. Concluding Remarks

Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guo, Y.; Gan, S.S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012, 35, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.R.; Jones, J.D.G. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef] [PubMed]
- Barna, B.; Fodor, J.; Harrach, B.; Pogány, M.; Király, Z. The janus face of reactive oxygen species in resistance and susceptibility of plants to necrotrophic and biotrophic pathogens. Plant Physiol. Biochem. 2012, 59, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Mengiste, T. Plant immunity to necrotrophs. Ann. Rev. Phytopathol. 2012, 50, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C.; Fitt, B.D.; Atkins, S.D.; Walters, D.R.; Daniell, T.J. Pathogenesis, parasitism and mutualism in the trophic space of microbe-plant interactions. Trends Microbiol. 2010, 18, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Gladders, P. Relevance of Verticillium wilt (Verticillium longisporum) in winter oilseed rape in the UK. HCGA Res. Rev. 2009, 72, 1–39. [Google Scholar]
- Abeles, F.B.; Morgan, P.W.; Saltveit, M.E., Jr. Ethylene in Plant Biology; Academic Press: San Diego, CA, USA, 2012. [Google Scholar]
- Grbić, V.; Bleecker, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602. [Google Scholar] [CrossRef]
- Jing, H.-C.; Schippers, J.H.; Hille, J.; Dijkwel, P.P. Ethylene-induced leaf senescence depends on age-related changes and old genes in Arabidopsis. J. Exp. Bot. 2005, 56, 2915–2923. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Geraats, B.P.; Linthorst, H.J. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006, 11, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Bailey, B.A. Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca. Phytopathology 1995, 85, 1250–1255. [Google Scholar] [CrossRef]
- Cristescu, S.M.; de Martinis, D.; te Lintel Hekkert, S.; Parker, D.H.; Harren, F.J. Ethylene production by Botrytis cinerea in vitro and in tomatoes. Appl. Environ. Microbiol. 2002, 68, 5342–5350. [Google Scholar] [CrossRef] [PubMed]
- Weingart, H.; Völksch, B. Ethylene production by Pseudomonas syringae pathovars in vitro and in planta. Appl. Environ. Microbiol. 1997, 63, 156–161. [Google Scholar] [PubMed]
- Penninckx, I.A.; Thomma, B.P.; Buchala, A.; Métraux, J.-P.; Broekaert, W.F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in arabidopsis. Plant Cell 1998, 10, 2103–2113. [Google Scholar] [CrossRef] [PubMed]
- Argueso, C.T.; Hansen, M.; Kieber, J.J. Regulation of ethylene biosynthesis. J. Plant Growth Regul. 2007, 26, 92–105. [Google Scholar] [CrossRef]
- Wang, K.L.-C.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, S131–S151. [Google Scholar] [PubMed]
- Hua, J.; Meyerowitz, E.M. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 1998, 94, 261–271. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Alonso, J.M. Ethylene signaling and response: Where different regulatory modules meet. Curr. Opin. Plant Biol. 2009, 12, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Yoon, G.M.; Shemansky, J.M.; Lin, D.Y.; Ying, Z.I.; Chang, J.; Garrett, W.M.; Kessenbrock, M.; Groth, G.; Tucker, M.L. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 19486–19491. [Google Scholar] [CrossRef] [PubMed]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y. Ethylene-induced stabilization of ETHYLENE INSENSITIVE 3 and EIN3-LIKE 1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhang, C.; Ji, Y.; Zhao, Q.; He, W.; An, F.; Jiang, L.; Guo, H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012, 22, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-D.; Cho, Y.; Sheen, J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009, 14, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Doubt, S.L. The Response of Plants to Illuminating Gas; University of Chicago: Chicago, IL, USA, 1917; Volume 227. [Google Scholar]
- Zimmerman, P.; Hitchcock, A.; Crocker, W. The effect of ethylene and illuminating gas on roses. Contrib. Boyce Thompson Inst. 1931, 3, 459–481. [Google Scholar]
- Jing, H.-C.; Anderson, L.; Sturre, M.J.; Hille, J.; Dijkwel, P.P. Arabidopsis cpr5 is a senescence-regulatory gene with pleiotropic functions as predicted by the evolutionary theory of senescence. J. Exp. Bot. 2007, 58, 3885–3894. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. ETHYLENE-INSENSITIVE 3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Köhler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/atNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Pré, M.; Atallah, M.; Champion, A.; de Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.-M.; To, T.K.; Li, W. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, J.; Deslandes, L.; Feng, D.X.; Balagué, C.; Marco, Y. Delayed symptom development in ein2–1, an Arabidopsis ethylene-insensitive mutant, in response to bacterial wilt caused by Ralstonia solanacearum. Phytopathology 2002, 92, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Staal, J.; Dixelius, C. Early responses in the Arabidopsis-Verticillium longisporum pathosystem are dependent on NDR1, JA- and ET-associated signals via cytosolic NPR1 and RFO1. Mol. Plant Microbe Interact. 2006, 19, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Pantelides, I.S.; Tjamos, S.E.; Paplomatas, E.J. Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae. Mol. Plant Pathol. 2010, 11, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Bleecker, A.B.; Kende, H. Ethylene: A gaseous signal molecule in plants. Ann. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wen, C.-K. Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes. Plant Physiol. 2012, 158, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernández-Calvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCF-COI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F. The Arabidopsis CORONATINE INSENSITIVE 1 protein is a jasmonate receptor. Plant Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef] [PubMed]
- Ueda, J.; Kato, J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 1980, 66, 246–249. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chang, C.; Tucker, M.L. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Wang, J.; Chua, L.; Jiang, D.; Peng, W.; Xie, D. The role of Arabidopsis RUBISCO ACTIVASE in jasmonate-induced leaf senescence. Plant Physiol. 2011, 155, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xing, D. Methyl jasmonate induces production of reactive oxygen species and alterations in mitochondrial dynamics that precede photosynthetic dysfunction and subsequent cell death. Plant Cell Physiol. 2008, 49, 1092–1111. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Zentgraf, U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 2007, 19, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; van Wees, S.C. Hormonal modulation of plant immunity. Ann. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Truman, W.; Bennett, M.H.; Kubigsteltig, I.; Turnbull, C.; Grant, M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. USA 2007, 104, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Ann. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Rivas-San Vicente, M.; Plasencia, J. Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 2011, 62, 3321–3338. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.; Mackerness, S.A.H.; Page, T.; John, C.F.; Murphy, A.M.; Carr, J.P.; Buchanan-Wollaston, V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 2000, 23, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Zentgraf, U.; Laun, T.; Miao, Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 2010, 89, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Rivas, S.; Roby, D. An essential role for salicylic acid in atMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett. 2006, 580, 3498–3504. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Ann. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.M. The complex regulation of senescence. Crit. Rev. Plant Sci. 2012, 31, 124–147. [Google Scholar] [CrossRef]
- Gazzarrini, S.; McCourt, P. Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 2001, 4, 387–391. [Google Scholar] [CrossRef]
- Vanhee, C.; Batoko, H. Autophagy involvement in responses to abscisic acid by plant cells. Autophagy 2011, 7, 655–656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Jiang, M.; Zhang, J.; Tan, M.; Hu, X. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 2006, 141, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Tsz-fung, F.C. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of start proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.; Tischer, S.V.; Wunschel, C.; Christmann, A.; Grill, E. Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc. Natl. Acad. Sci. USA 2014, 111, 5741–5746. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, C. Signal transduction in leaf senescence. Plant Mol. Biol. 2013, 82, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Gepstein, S.; Thimann, K.V. Changes in the abscisic acid content of oat leaves during senescence. Proc. Natl. Acad. Sci. USA 1980, 77, 2050–2053. [Google Scholar] [CrossRef] [PubMed]
- Weaver, L.M.; Gan, S.; Quirino, B.; Amasino, R.M. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 1998, 37, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Nam, H.G.; Lin, J.F.; Wu, S.H.; Swidzinski, J.; Ishizaki, K. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.C.; Hong, S.W.; Whang, S.S.; Lim, P.O.; Nam, H.G.; Koo, J.C. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 2011, 52, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Ni, D.-A.; Ruan, Y.-L. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 2009, 21, 2072–2089. [Google Scholar] [CrossRef] [PubMed]
- Balibrea Lara, M.E.; Gonzalez Garcia, M.-C.; Fatima, T.; Ehneß, R.; Lee, T.K.; Proels, R.; Tanner, W.; Roitsch, T. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 2004, 16, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Godt, D.E.; Roitsch, T. Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiol. 1997, 115, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Garapati, P.; Xue, G.-P.; Munné-Bosch, S.; Balazadeh, S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-D.; Seo, P.J.; Yoon, H.-K.; Park, C.-M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef] [PubMed]
- Raab, S.; Drechsel, G.; Zarepour, M.; Hartung, W.; Koshiba, T.; Bittner, F.; Hoth, S. Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J. 2009, 59, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Vogelmann, K.; Drechsel, G.; Bergler, J.; Subert, C.; Philippar, K.; Soll, J.; Engelmann, J.C.; Engelsdorf, T.; Voll, L.M.; Hoth, S. Early senescence and cell death in Arabidopsis saul1 mutants involves the PAD4-dependent salicylic acid pathway. Plant Physiol. 2012, 159, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef] [PubMed]
- Audenaert, K.; de Meyer, G.B.; Höfte, M.M. Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 2002, 128, 491–501. [Google Scholar] [CrossRef] [PubMed]
- De Torres-Zabala, M.; Truman, W.; Bennett, M.H.; Lafforgue, G.; Mansfield, J.W.; Rodriguez Egea, P.; Bögre, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [CrossRef]
- Häffner, E.; Karlovsky, P.; Splivallo, R.; Traczewska, A.; Diederichsen, E. ERECTA, salicylic acid, abscisic acid and jasmonic acid modulate quantitative disease resistance of Arabidopsis thaliana to Vverticillium longisporum. BMC Plant Biol. 2014, 14, 85. [Google Scholar] [CrossRef] [PubMed]
- Ulferts, S.; Delventhal, R.; Splivallo, R.; Karlovsky, P.; Schaffrath, U. Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae. BMC Plant Biol. 2015, 15, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Torres-Zabala, M.; Bennett, M.H.; Truman, W.H.; Grant, M.R. Antagonism between salicylic and abscisic acid reflects early host–pathogen conflict and moulds plant defence responses. Plant J. 2009, 59, 375–386. [Google Scholar] [PubMed]
- Jiang, C.-J.; Shimono, M.; Sugano, S.; Kojima, M.; Yazawa, K.; Yoshida, R.; Inoue, H.; Hayashi, N.; Sakakibara, H.; Takatsuji, H. Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 2010, 23, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Adie, B.; Pérez-Pérez, J.; Pérez-Pérez, M.; Godoy, M.; Sánchez-Serrano, J.-J.; Schmelz, E.; Solano, R. ABA is an essential signal for plant resistance to pathogens affecting ja biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 2007, 19, 1665–1681. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via atMPK6-coupled signaling in Arabidopsis. Plant J. 2008, 54, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Jibran, R.; Hunter, D.A.; Dijkwel, P.P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gou, X.; Yuan, T.; Lin, H.; Asami, T.; Yoshida, S.; Russell, S.D.; Li, J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 2007, 17, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Nakashita, H.; Yasuda, M.; Nitta, T.; Asami, T.; Fujioka, S.; Arai, Y.; Sekimata, K.; Takatsuto, S.; Yamaguchi, I.; Yoshida, S. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003, 33, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Divi, U.K.; Rahman, T.; Krishna, P. Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 2010, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Krishna, P. Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 2003, 22, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, K.; Wei, J.; Ma, Q.; Wang, B.; Yu, D. Gibberellin retards chlorophyll degradation during senescence of Paris polyphylla. Biol. Plant. 2010, 54, 395–399. [Google Scholar] [CrossRef]
- Yu, K.; Wang, Y.; Wei, J.; Ma, Q.; Yu, D.; Li, J. Improving rhizome yield and quality of Paris polyphylla through gibberellic acid-induced retardation of senescence of aerial parts. Plant Signal. Behav. 2009, 4, 413–415. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef] [PubMed]
- Merewitz, E.B.; Gianfagna, T.; Huang, B. Photosynthesis, water use, and root viability under water stress as affected by expression of SAG12-IPT controlling cytokinin synthesis in Agrostis stolonifera. J. Exp. Bot. 2011, 62, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Schmülling, T.; Werner, T.; Riefler, M.; Krupková, E.; Bartrina y Manns, I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003, 116, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [PubMed]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Slovin, J.P.; Hendrickson, A.M. Two genetically discrete pathways convert tryptophan to auxin: More redundancy in auxin biosynthesis. Trends Plant Sci. 2003, 8, 197–199. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Noh, Y.-S.; Amasino, R.M. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 1999, 41, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Kim, J-I.; Murphy, A.S.; Baek, D.; Lee, S.-W.; Yun, D.-J.; Bressan, R.A.; Narasimhan, M.L. YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2011. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Lee, I.C.; Kim, J.; Kim, H.J.; Ryu, J.S.; Woo, H.R.; Nam, H.G. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 2010. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liang, G.; Yang, S.; Yu, D. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid–and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef] [PubMed]
- Quirino, B.F.; Normanly, J.; Amasino, R.M. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. 1999, 40, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flügge, U.-I.; Kunze, R. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Meng, T.; Li, P.; Yu, Y.; Cui, Y.; Wang, Y.; Gong, Q.; Wang, N.N. A soybean dual-specificity kinase, gmSARK, and its Arabidopsis homolog, atSARK, regulate leaf senescence through synergistic actions of auxin and ethylene. Plant Physiol. 2011, 157, 2131–2153. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, W.; Gan, S.-S. Saur36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol. 2013, 161, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Choi, D.; Lee, S.; Ryu, C.-M.; Hwang, I. Cytokinins and plant immunity: Old foes or new friends? Trends Plant Sci. 2011, 16, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. Linking development to defense: Auxin in plant–pathogen interactions. Trends Plant Sci. 2009, 14, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-P. The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Curr. Biol. 2011, 21, R338–R345. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 2012, 17, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; He, X.; Luo, X.; Xu, L.; Liu, L.; Min, L.; Jin, L.; Zhu, L.; Zhang, X. Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE-ZIM-DOMAIN1 expression. Plant Physiol. 2014, 166, 2179–2194. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Lee, L.Y.C.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.-T.; Sun, T.-P.; Li, J.; Deng, X.-W. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Bari, R.; Achard, P.; Lisón, P.; Nemri, A.; Harberd, N.P.; Jones, J.D. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 2008, 18, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, M.J.; Terrile, M.C.; Casalongué, C.A. Auxin and salicylic acid signalings counteract the regulation of adaptive responses to stress. Plant Signal. Behav. 2011, 6, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Park, J.-Y.; Kim, Y.-S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.-Y.; Kim, J.; Lee, Y.-H.; Park, C.-M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Cao, Y.; Huang, L.; Zhao, J.; Xu, C.; Li, X.; Wang, S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate-and jasmonate-independent basal immunity in rice. Plant Cell 2008, 20, 228–240. [Google Scholar] [CrossRef] [PubMed]
- González-Lamothe, R.; El Oirdi, M.; Brisson, N.; Bouarab, K. The conjugated auxin indole-3-acetic acid-aspartic acid promotes plant disease development. Plant Cell 2012, 24, 762–777. [Google Scholar] [CrossRef] [PubMed]
- Llorente, F.; Muskett, P.; Sánchez-Vallet, A.; López, G.; Ramos, B.; Sánchez-Rodríguez, C.; Jordá, L.; Parker, J.; Molina, A. Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol. Plant 2008, 1, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Pan, J.; Peng, W.; Genschik, P.; Hobbie, L.; Hellmann, H.; Estelle, M.; Gao, B.; Peng, J.; Sun, C. Point mutations in Arabidopsis CULLIN1 reveal its essential role in jasmonate response. Plant J. 2005, 42, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Kidd, B.N.; Kadoo, N.Y.; Dombrecht, B.; Tekeoglu, M.; Gardiner, D.M.; Thatcher, L.F.; Aitken, E.A.; Schenk, P.M.; Manners, J.M.; Kazan, K. Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Mol. Plant-Microbe Interact. 2011, 24, 733–748. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Riaño-Pachón, D.; Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 2008, 10, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed]
- Kiełbowicz-Matuk, A. Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 2012, 185, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Amornsiripanitch, N.; Dong, X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens 2006, 2, e123. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Reddy, M.; Chikara, J. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. Targets of atWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-C.; Lai, Z.; Fan, B.; Chen, Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, C.; Fan, B.; Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY 40, and WRKY 60 transcription factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Yan, L.; Liu, Z.-Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.-Q.; Wang, X.-F.; Du, S.-Y.; Jiang, T. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Li, D.; Wang, F.; Yu, D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, E1963–E1971. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Ann. Rev. Plant Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wang, Y.; Zheng, X.; Jia, Q.; Zhao, J.; Bai, F.; Hong, Y.; Liu, Y. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 2015, 27, 1316–1331. [Google Scholar] [CrossRef] [PubMed]
- Van Breusegem, F.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006, 141, 384–390. [Google Scholar]
- De Gara, L.; de Pinto, M.C.; Tommasi, F. The antioxidant systems vis-à-vis reactive oxygen species during plant-pathogen interaction. Plant Physiol. Biochem. 2003, 41, 863–870. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Dangl, J.L. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Pathogen-induced, NADPH oxidase–derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 2005, 37, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Rodriguez, R.; Tran, A.; Hoang, H.; de los Santos, D.; Brown, S.; Vellanoweth, R.L. The developmental transition to flowering represses ascorbate peroxidase activity and induces enzymatic lipid peroxidation in leaf tissue in Arabidopsis thaliana. Plant Sci. 2000, 158, 115–127. [Google Scholar] [CrossRef]
- Zimmermann, P.; Zentgraf, U. The correlation between oxidative stress and leaf senescence during plant development. Cell. Mol. Biol. Letters 2005, 10, 515. [Google Scholar]
- Navabpour, S.; Morris, K.; Allen, R.; Harrison, E.; Soheila, A.; Buchanan-Wollaston, V. Expression of senescence-enhanced genes in response to oxidative stress. J. Exp. Bot. 2003, 54, 2285–2292. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.-I.; Asensi-Fabado, M.A.; Munné-Bosch, S.; Antonio, C.; Tohge, T. JUNGBRUNNEN1, a reactive oxygen species–responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Wu, A.; Mueller-Roeber, B. Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal. Behav. 2010, 5, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.-P.; Mueller-Roeber, B. ORS1, an H2 O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2010, 138, 414–429. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Saga, H.; Ogawa, T.; Kai, K.; Suzuki, H.; Ogata, Y.; Sakurai, N.; Shibata, D.; Ohta, D. Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol. Plant Microbe Interact. 2012, 25, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Blume, B.; Nürnberger, T.; Nass, N.; Scheel, D. Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell 2000, 12, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.J.; Yun, B.W.; Loake, G.J. Oxidative burst and cognate redox signalling reported by luciferase imaging: Identification of a signal network that functions independently of ethylene, SA and Me-JA but is dependent on MAPKK activity. Plant J. 2000, 24, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase atRBOHD and atRBOHF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Li, G.J.; Yang, K.Y.; Mao, G.; Wang, R.; Liu, Y.; Zhang, S. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010, 64, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Blomster, T.; Salojärvi, J.; Sipari, N.; Brosché, M.; Ahlfors, R.; Keinänen, M.; Overmyer, K.; Kangasjärvi, J. Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in Arabidopsis. Plant Physiol. 2011, 157, 1866–1883. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.; Reynolds, A.; Hancock, J.; Neill, S. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem. J. 1998, 330, 115–120. [Google Scholar] [PubMed]
- Hennin, C.; Diederichsen, E.; Höfte, M. Local and systemic resistance to fungal pathogens triggered by an AVR9-mediated hypersensitive response in tomato and oilseed rape carrying the Cf-9 resistance gene. Physiol. Mol. Plant Pathol. 2001, 59, 287–295. [Google Scholar] [CrossRef]
- Harrach, B.D.; Fodor, J.; Pogány, M.; Preuss, J.; Barna, B. Antioxidant, ethylene and membrane leakage responses to powdery mildew infection of near-isogenic barley lines with various types of resistance. Eur. J. Plant Pathol. 2008, 121, 21–33. [Google Scholar] [CrossRef]
- Williams, B.; Kabbage, M.; Kim, H.-J.; Britt, R.; Dickman, M.B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathogens 2011, 7, e1002107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avila-Ospina, L.; Moison, M.; Yoshimoto, K.; Masclaux-Daubresse, C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for self-eating in plant cells. Ann. Rev. Plant Biol. 2012, 63, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011, 66, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Czymmek, K.; Talloczy, Z.; Levine, B.; Dinesh-Kumar, S. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Jikumaru, Y.; Kamiya, Y.; Kusano, M.; Consonni, C.; Panstruga, R.; Ohsumi, Y.; Shirasu, K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 2009, 21, 2914–2927. [Google Scholar] [CrossRef] [PubMed]
- Hofius, D.; Munch, D.; Bressendorff, S.; Mundy, J.; Petersen, M. Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 2011, 18, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Hulbert, S.; Bai, J.; Fellers, J.; Pacheco, M.; Bowden, R. Gene expression patterns in near isogenic lines for wheat rust resistance gene Lr34/Yr18. Phytopathology 2007, 97, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Uauy, C.; Distelfeld, A.; Blechl, A.; Epstein, L.; Chen, X.; Sela, H.; Fahima, T.; Dubcovsky, J. A kinase-start gene confers temperature-dependent resistance to wheat stripe rust. Science 2009, 323, 1357–1360. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.-Y.; Li, K.; Wu, K.; Wang, X.; Lin, H.; Cantu, D.; Uauy, C.; Dobon-Alonso, A.; Midorikawa, T.; Inoue, K. Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell 2015. [Google Scholar] [CrossRef] [PubMed]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Piffanelli, P.; Zhou, F.; Casais, C.; Orme, J.; Jarosch, B.; Schaffrath, U.; Collins, N.C.; Panstruga, R.; Schulze-Lefert, P. The barley Mlo modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002, 129, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Jarosch, B.; Collins, N.C.; Zellerhoff, N.; Schaffrath, U. RAR1, ROR1, and the actin cytoskeleton contribute to basal resistance to Magnaporthe grisea in barley. Mol. Plant Microbe Interact. 2005, 18, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Muchero, W.; Ehlers, J.D.; Close, T.J.; Roberts, P.A. Genic SNP markers and legume synteny reveal candidate genes underlying QTL for Macrophomina phaseolina resistance and maturity in cowpea [Vigna unguiculata (L) Walp.]. BMC Genomics 2011, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Mah, K.M.; Uppalapati, S.R.; Tang, Y.; Allen, S.; Shuai, B. Gene expression profiling of Macrophomina phaseolina infected Medicago truncatula roots reveals a role for auxin in plant tolerance against the charcoal rot pathogen. Physiol. Mol. Plant Pathol. 2012, 79, 21–30. [Google Scholar] [CrossRef]
- Blanco-Ulate, B.; Vincenti, E.; Powell, A.L.; Cantu, D. Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Front. Plant Sci. 2013, 4, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Hu, Q.; Johansson, A.; Dixelius, C. Verticillium longisporum and V. dahliae: Infection and disease in Brassica napus. Plant Pathol. 2006, 55, 137–144. [Google Scholar] [CrossRef]
- Häffner, E.; Karlovsky, P.; Diederichsen, E. Genetic and environmental control of the Verticillium syndrome in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 235. [Google Scholar] [CrossRef] [PubMed]
- Veronese, P.; Narasimhan, M.L.; Stevenson, R.A.; Zhu, J.-K.; Weller, S.C.; Subbarao, K.V.; Bressan, R.A. Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J. 2003, 35, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Smalle, J.; Va, M.; Montagu, N.; Inzé, D. Oxidative stress tolerance and longevity in Arabidopsis: The late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998, 14, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.L.; Madden, L.V. Introduction to Plant Disease Epidemiology; John Wiley and Sons: New York, NY, USA, 1990. [Google Scholar]
- Robert-Seilaniantz, A.; Navarro, L.; Bari, R.; Jones, J.D. Pathological hormone imbalances. Curr. Opin. Plant Biol. 2007, 10, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Agnew, J.L.; Cohen, J.D.; He, P.; Shan, L.; Sheen, J.; Kunkel, B.N. Pseudomonas syringae type III effector avrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA 2007, 104, 20131–20136. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Müller, J.; Prinsen, E.; Rolfe, S.A.; Scholes, J.D. Metabolism and plant hormone action during clubroot disease. J. Plant Growth Regul. 2009, 28, 229–244. [Google Scholar] [CrossRef]
- Dekhuijzen, H. The occurrence of free and bound cytokinins in plasmodia of Plasmodiophora brassicae isolated from tissue cultures of clubroots. Plant Cell Rep. 1981, 1, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Siemens, J.; Keller, I.; Sarx, J.; Kunz, S.; Schuller, A.; Nagel, W.; Schmülling, T.; Parniske, M.; Ludwig-Müller, J. Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant Microbe Interact. 2006, 19, 480–494. [Google Scholar] [CrossRef] [PubMed]
- Walters, D.R.; McRoberts, N.; Fitt, B.D. Are green islands red herrings? Significance of green islands in plant interactions with pathogens and pests. Biol. Rev. 2008, 83, 79–102. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Nürnberger, T.; Joosten, M.H. Of PAMPS and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Ralhan, A.; Schöttle, S.; Thurow, C.; Iven, T.; Feussner, I.; Polle, A.; Gatz, C. The vascular pathogen Verticillium longisporum requires a jasmonic acid-independent COI1 function in roots to elicit disease symptoms in Arabidopsis shoots. Plant Physiol. 2012, 159, 1192–1203. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, L.F.; Manners, J.M.; Kazan, K. Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 2009, 58, 927–939. [Google Scholar] [CrossRef] [PubMed]
- El Oirdi, M.; El Rahman, T.A.; Rigano, L.; El Hadrami, A.; Rodriguez, M.C.; Daayf, F.; Vojnov, A.; Bouarab, K. Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 2011, 23, 2405–2421. [Google Scholar] [CrossRef] [PubMed]
- Ottmann, C.; Luberacki, B.; Küfner, I.; Koch, W.; Brunner, F.; Weyand, M.; Mattinen, L.; Pirhonen, M.; Anderluh, G.; Seitz, H.U. A common toxin fold mediates microbial attack and plant defense. Proc. Natl. Acad. Sci. USA 2009, 106, 10359–10364. [Google Scholar] [CrossRef] [PubMed]
- Barau, J.; Grandis, A.; de Andrade Carvalho, V.M.; Teixeira, G.S.; Zaparoli, G.H.A.; do Rio, M.C.S.; Rincones, J.; Buckeridge, M.S.; Pereira, G.A.G. Apoplastic and intracellular plant sugars regulate developmental transitions in witches’ broom disease of cacao. J. Exp. Bot. 2014. [Google Scholar] [CrossRef] [PubMed]
- Oliver, R.P.; Solomon, P.S. New developments in pathogenicity and virulence of necrotrophs. Curr. Opin. Plant Biol. 2010, 13, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Faris, J.D.; Oliver, R.P.; Tan, K.-C.; Solomon, P.S.; McDonald, M.C.; McDonald, B.A.; Nunez, A.; Lu, S.; Rasmussen, J.B. SnTOX3 acts in effector triggered susceptibility to induce disease on wheat carrying the snn3 gene. PLoS pathogens 2009, 5, e1000581. [Google Scholar] [CrossRef] [PubMed]
- Hornig, H. Krankhafte Abreife—Spätschäden durch Phoma lingam und/oder Verticillium dahliae. Raps 1986, 4, 83–85. [Google Scholar]
- Eynck, C.; Koopmann, B.; Karlovsky, P.; von Tiedemann, A. Internal resistance in winter oilseed rape inhibits systemic spread of the vascular pathogen Verticillium longisporum. Phytopathology 2009, 99, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- NCBI. GEO Gene Expression Omnibus. Available online: http://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/geo/query/acc.cgi?acc=GSE70021 (accessed on 8 July 2015).
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. Map kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Craigon, D.J.; James, N.; Okyere, J.; Higgins, J.; Jotham, J.; May, S. Nascarrays: A repository for microarray data generated by NASC’s transcriptomics service. Nucleic Acids Res. 2004, 32, D575–D577. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V., Dudoit, S., Irizarry, R., Huber, W., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Carvalho, B. pd.aragene.1.1.st: Platform Design info for Affymetrix Aragene-1_1-st., R package version 3.8.0. Available online: http://www.bioconductor.org/packages/release/data/annotation/html/pd.aragene.1.1.st.html (accessed on 7 July 2015).
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Pousada, R.A.; de Rycke, R.; Dedonder, A.; van Caeneghem, W.; Engler, G.; van Montagu, M.; van der Straeten, D. The Arabidopsis 1-aminocyclopropane-1-carboxylate synthase gene 1 is expressed during early development. Plant Cell 1993, 5, 897–911. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Sánchez, L.; Anderson, J.P.; Young, J.; Singh, K.B. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2007, 143, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Harvey Millar, A.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009, 58, 53–68. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Häffner, E.; Konietzki, S.; Diederichsen, E. Keeping Control: The Role of Senescence and Development in Plant Pathogenesis and Defense. Plants 2015, 4, 449-488. https://0-doi-org.brum.beds.ac.uk/10.3390/plants4030449
Häffner E, Konietzki S, Diederichsen E. Keeping Control: The Role of Senescence and Development in Plant Pathogenesis and Defense. Plants. 2015; 4(3):449-488. https://0-doi-org.brum.beds.ac.uk/10.3390/plants4030449
Chicago/Turabian StyleHäffner, Eva, Sandra Konietzki, and Elke Diederichsen. 2015. "Keeping Control: The Role of Senescence and Development in Plant Pathogenesis and Defense" Plants 4, no. 3: 449-488. https://0-doi-org.brum.beds.ac.uk/10.3390/plants4030449





