Applications of Electronic-Nose Technologies for Noninvasive Early Detection of Plant, Animal and Human Diseases
Abstract
:1. Introduction
2. Biomarker Metabolite Electronic-Nose Signatures
3. Electronic-Nose Detection of Biotic Diseases
3.1. Plant Disease Detection
3.2. Animal Disease Detection
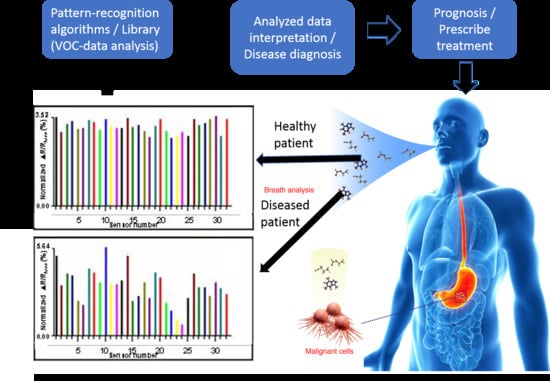
3.3. Human Disease Detection
Volatile Organic Compound -Biomarkers of Human Diseases
4. Future Electronic-Nose Technologies for Improved Disease Diagnostics
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Bos, L.D.; Sterk, P.J.; Schultz, M.J. Volatile metabolites of pathogens: A systematic review. PLoS Pathog. 2013, 9, e1003311. [Google Scholar] [CrossRef] [PubMed]
- Santini, G.; Mores, N.; Penas, A.; Capuano, R.; Mondino, C.; Trové, A.; Macagno, F.; Zini, G.; Cattani, P.; Martinelli, E.; et al. Electronic nose and exhaled breath NMR-based metabolomics applications in airways disease. Curr. Top. Med. Chem. 2016, 16, 1610–1630. [Google Scholar] [CrossRef] [PubMed]
- Spaněl, P.; Smith, D. Progress in SIFT-MS: Breath analysis and other applications. Mass Spectrom. Rev. 2011, 30, 236–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sahay, P. Breath analysis using laser spectroscopic techniques: Breath biomarkers, spectral fingerprints, and detection limits. Sensors 2009, 9, 8230–8262. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D.; Baietto, M. Applications and advances in electronic-nose technologies. Sensors 2009, 9, 5099–5148. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Recent progress in the design and clinical development of electronic-nose technologies. Nanobiosens. Dis. Diagn. 2016, 5, 15–27. [Google Scholar] [CrossRef]
- Wilson, A.D. Biomarker metabolite signatures pave the way for electronic-nose applications in early clinical disease diagnoses. Curr. Metabolomics 2017, 5, 90–101. [Google Scholar] [CrossRef]
- Wilson, A.D. Electronic-nose devices—Potential for noninvasive early disease-detection applications. Ann. Clin. Case Rep. 2017, 2, 1401. Available online: http://www.anncaserep.com/full-text/accr-v2-id1401.php (accessed on 14 July 2017).
- Cellini, A.; Blasioli, S.; Biondi, E.; Bertaccini, A.; Braschi, I.; Spinelli, F. Potential applications and limitations of electronic nose devices for plant disease diagnosis. Sensors 2017, 17, 2596. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D.; Lester, D.G.; Oberle, C.S. Development of conductive polymer analysis for the rapid detection and identification of phytopathogenic microbes. Phytopathology 2004, 94, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Casalinuovo, I.A.; Di Pierro, D.; Coletta, M.; Di Francesco, P. Application of electronic noses for disease diagnosis and food spoilage detection. Sensors 2006, 6, 1428–1439. [Google Scholar] [CrossRef]
- Wilson, A.D.; Baietto, M. Advances in electronic-nose technologies developed for biomedical applications. Sensors 2011, 11, 1105–1176. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Diverse applications of electronic nose technologies in agriculture and forestry. Sensors 2013, 13, 2295–2348. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.A.; Shevade, A.V.; Taylor, C.J.; Homer, M.L.; Jewell, A.D.; Kisor, A.; Manatt, K.S.; Yen, S.P.S.; Blanco, M.; Goddard, W.A. Expanding the Capabilities of the JPL Electronic Nose for an International Space Station Technology Demonstration; SAE Technical Paper 2006-01-2179; Jet Propulsion Laboratory, National Aeronautics and Space Administration: Pasadena, CA, USA, 2006.
- Shevade, A.V.; Homer, M.L.; Zhou, H.; Jewell, A.D.; Kisor, A.K.; Manatt, K.S.; Torres, J.; Soler, J.; Yen, S.P.S.; Ryan, M.A.; et al. Development of the Third Generation JPL Electronic Nose for International Space Station Technology Demonstration; SAE Technical Paper 2007-01-3149; Jet Propulsion Laboratory, California Institute of Technology: Pasadena, CA, USA, 2007. [Google Scholar]
- Ryan, M.A.; Manatt, K.S.; Gluck, S.E.; Shevade, A.V.; Kisor, A.K.; Zhou, H.; Lara, L.M.; Homer, M.L. Operation of Third Generation JPL Electronic Nose on the International Space Station; SAE Technical Paper 2009-01-2522; Jet Propulsion Laboratory, California Institute of Technology: Pasadena, CA, USA, 2009. [Google Scholar]
- Kateb, B.; Ryan, M.A.; Homer, M.L.; Lara, L.M.; Yin, Y.; Higa, K.; Chen, M.Y. Sniffing out cancer using the JPL electronic nose: A pilot study of a novel approach to detection and differentiation of brain cancer. NeuroImage 2009, 47, T5–T9. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Review of electronic-nose technologies and algorithms to detect hazardous chemicals in the environment. Procedia Technol. 2012, 1, 453–463. [Google Scholar] [CrossRef]
- Wilson, A.D.; Lester, D.G.; Oberle, C.S. Application of conductive polymer analysis for wood and woody plant identifications. For. Ecol. Manag. 2005, 209, 207–224. [Google Scholar] [CrossRef]
- Wilson, A.D. Future applications of electronic‑nose technologies in healthcare and biomedicine. In Wide Spectra of Quality Control; Akyar, I., Ed.; InTech Publishing: Rijeka, Croatia, 2011; ISBN 978-953-307-683-6. [Google Scholar]
- Wilson, A.D. Finding Aroma Clues in the Human Breath to Diagnose Diseases. Atlas of Science 29 February 2016. Available online: http://atlasofscience.org/finding-aroma-clues-in-the-human-breath-to-diagnose-diseases (accessed on 20 August 2018).
- Wilson, A.D. Identification of insecticide residues with a conducting-polymer electronic nose. Chem. Sens. 2014, 4, 1–10. [Google Scholar]
- Wilson, A.D. Fungicide residue identification and discrimination using a conducting polymer electronic-nose. In Proceedings of the IV International Conference on Sensor Device Technologies and Applications, Barcelona, Spain, 25–31 August 2013; Yurish, S., Chilibon, I., Carvalho, V., Gervais-Ducouret, S., Eds.; Xpert Publishing Services: Wilmington, DE, USA, 2013; pp. 116–121. [Google Scholar]
- Wilson, A.D. Identification and discrimination of herbicide residues using a conducting polymer electronic nose. In Proceedings of the VII International Conference on Sensor Device Technologies and Applications, Nice, France, 24–28 July 2016; International Academy, Research, and Industry Association (IARIA): Wilmington, DE, USA, 2016; pp. 4–7. [Google Scholar]
- Baldwin, E.A.; Bai, J.; Plotto, A.; Dea, S. Electronic noses and tongues: Application for the food and pharmaceutical industries. Sensors 2011, 11, 4744–4766. [Google Scholar] [CrossRef] [PubMed]
- Baietto, M.; Wilson, A.D. Electronic-nose applications for fruit identification, ripeness and quality grading. Sensors 2015, 15, 899–931. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Electronic-nose applications in forensic science and for analysis of volatile biomarkers in the human breath. J. Forensic Sci. Criminol. 2014, 1, S103. [Google Scholar] [CrossRef]
- Wilson, A.D. Theoretical and practical considerations for teaching diagnostic electronic-nose technologies to clinical laboratory technicians. Proc. Soc. Behav. Sci. 2012, 31, 262–274. [Google Scholar] [CrossRef]
- Wilson, A.D. Advanced methods for teaching electronic-nose technologies to diagnosticians and clinical laboratory technicians. Proc. Soc. Behav. Sci. 2012, 46, 4544–4554. [Google Scholar] [CrossRef]
- Li, H.; Tang, H.; Wang, Y. Advances in metabonomics on infectious diseases. Curr. Metabolomics 2013, 1, 318–334. [Google Scholar] [CrossRef]
- Emwas, A.M.; Salek, R.M. NMR-based metabolomics in human disease diagnosis: Applications, limitations, and recommendations. Metabolomics 2013, 9, 1048–1072. [Google Scholar] [CrossRef]
- Wilson, A.D. Advances in electronic-nose technologies for the detection of volatile biomarker metabolites in the human breath. Metabolites 2015, 5, 140–163. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, F.; Cellini, A.; Vanneste, J.L.; Rodriguez-Estrada, M.T.; Costa, G.; Savioli, S.; Harren, F.J.M.; Cristescu, S.M. Emission of volatile compounds by Erwinia amylovora biological activity in vitro and possible exploitation for bacterial identification. Trees 2012, 26, 141–152. [Google Scholar] [CrossRef]
- Cellini, A.; Biondi, E.; Blasioli, S.; Rocchi, L.; Farneti, B.; Braschi, I.; Savioli, S.; Rodriguez-Estrada, M.T.; Biasioli, F.; Spinelli, F. Early detection of bacterial diseases in apple plants by analysis of volatile organic compounds profiles and use of electronic nose. Ann. Appl. Biol. 2016, 168, 409–420. [Google Scholar] [CrossRef]
- Thelen, J.; Harbinson, J.; Jansen, R.; Van Straten, G.; Posthumus, M.A.; Woltering, E.J.; Bouwmeester, H.J. The sesquiterpene α-copaene is induced in tomato leaves infected by Botrytis cinerea. J. Plant Interact. 2005, 1, 163–170. [Google Scholar] [CrossRef]
- Laothawornkitkul, J.; Moore, J.P.; Taylor, J.E.; Possell, M.; Gibson, T.D.; Hewett, C.N.; Paul, N.D. Discrimination of plant volatile signatures by an electronic nose: A potential technology for plant pest and disease monitoring. Environ. Sci. Technol. 2008, 42, 8433–8439. [Google Scholar] [CrossRef] [PubMed]
- Peled, N.; Koslow, M.; Ryan, J.; Haick, H. Detection of volatile organic compounds in cattle naturally infected with Mycobacterium bovis. Sens. Actuators B 2012, 171–172, 588–594. [Google Scholar] [CrossRef]
- Van den Velde, S.; van Steenberghe, D.; van Hee, P.; Quirynen, M. Detection of odorous compounds in breath. J. Dent. Res. 2009, 88, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Okell, C.C.; Elliott, S.D. Bacteraemia and oral sepsis with special reference to the aetiology of subacute endocarditis. Lancet 1935, 2, 869–872. [Google Scholar] [CrossRef]
- Drangsholt, M.T. A new causal model of dental diseases associated with endocarditis. Ann. Periodontol. 1998, 3, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Lacassin, F.; Hoen, B.; Leport, C.; Selton-Suty, C.; Delahaye, F.; Goulet, V.; Etienne, J.; Briançon, S. Procedures associated with infective endocarditis in adults: A case-control study. Eur. Heart J. 1995, 16, 1968–1974. [Google Scholar] [CrossRef] [PubMed]
- Balint, B.; Kharitonov, S.A.; Hanazawa, T.; Donnelly, L.E.; Shah, P.L.; Hodson, M.E.; Barnes, P.J. Increased nitrotyrosine in exhaled breath condensate in cystic fibrosis. Eur. Respir. J. 2001, 17, 1201–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, A.D. Recent applications of electronic-nose technologies for the noninvasive early diagnosis of gastrointestinal diseases. Proceedings 2018, 2, 147. [Google Scholar] [CrossRef]
- Wilson, A.D. Application of electronic-nose technologies and VOC-biomarkers for the noninvasive early diagnosis of gastrointestinal diseases. Sensors 2018, 18, 2613. [Google Scholar] [CrossRef] [PubMed]
- Taware, R.; Taunk, K.; Pereira, JA.; Dhakne, R.; Kannan, N.; Soneji, D.; Câmara, J.; Nagara, H.A.; Rapole, S. Investigation of urinary volatomic alterations in head and neck cancer: A non-invasive approach towards diagnosis and prognosis. Metabolomics 2017, 13, 111. [Google Scholar] [CrossRef]
- Paredes, R.M.G.; Pinto, C.G.; Pavón, J.L.P.; Cordero, B.M. Headspace-gas chromatography-mass spectrometry for the rapid determination of possible biomarkers in urine samples. Anal. Methods 2017, 9, 5784–5790. [Google Scholar] [CrossRef]
- Bax, C.; Taverna, G.; Eusebio, L.; Sironi, S.; Grizzi, F.; Guazzoni, G.; Capelli, L. Innovative diagnostic methods for early prostate cancer detection through urine analysis: A review. Cancers 2018, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Van Oort, P.; Povoa, P.; Schnabel, R.; Dark, P.M.; Artigas, A.; Bergmans, D.C.J.-J.; Felton, T.W.; Coelho, L.; Schultz, M.J.; Fowler, S.J.; et al. The potential role of exhaled breath analysis in the diagnostic process of pneumonia—A systematic review. J. Breath Res. 2018, 12, 024001. [Google Scholar] [CrossRef] [PubMed]
- Bos, L.D.; Sterk, P.J.; Fowler, S.J. Breathomics in the setting of asthma and chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2016, 138, 970–976. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, R.; Fijten, R.; Smolinska, A.; Dallinga, J.; Boumans, M.-L.; Stobberingh, E.; Boots, A.; Roekaerts, P.; Bergmans, D.; van Schooten, F.J. Analysis of volatile organic compounds in exhaled breath to diagnose ventilator-associated pneumonia. Sci. Rep. 2015, 5, 17179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, S.J.; Basanta-Sanchez, M.; Xu, Y.; Goodacre, R.; Dark, P.M. Surveillance for lower airway pathogens in mechanically ventilated patients by metabolomic analysis of exhaled breath: A case-control study. Thorax 2015, 70, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Van Oort, P.M.P.; De Bruin, S.; Weda, H.; Knobel, H.H.; Schultz, M.J.; Bos, L.D. Exhaled breath metabolomics for the diagnosis of pneumonia in intubated and mechanically-ventilated intensive care unit (ICU)-patients. Int. J. Mol. Sci. 2017, 18, 449. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.R.; Wang, Y.L. Metabonomics: A revolution in progress. Prog. Biochem. Biophys. 2006, 33, 401–417. [Google Scholar]
- Pardo, M.; Sberveglieri, G. Electronic olfactory systems based on metal oxide semiconductor sensor arrays. MRS Bull. 2004, 29, 703–708. [Google Scholar] [CrossRef]
- Loutfi, A.; Coradeschi, S.; Mani, G.K.; Shankar, P.; Rayappan, J.B.B. Electronic noses for food quality: A review. J. Food Eng. 2015, 144, 103–111. [Google Scholar] [CrossRef]
- Peris, M.; Escuder-Gilabert, L. A 21st century technique for food control: Electronic noses. Anal. Chim. Acta 2009, 638, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D. Application of a conductive polymer electronic-nose device to identify aged woody samples. In Proceedings of the 3rd International IARIA Conference on Sensor Device Technologies and Applications, Rome, Italy, 19–24 August 2012; Xpert Publishing Services: Wilmington, DE, USA, 2012; pp. 77–82, ISBN 978-1-61208-208-0. [Google Scholar]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Ray, M.; Ray, A.; Dash, S.; Mishra, A.; Achary, K.G.; Nayak, S.; Singh, S. Fungal disease detection in plants: Traditional assays, novel diagnostic techniques and biosensors. Biosens. Bioelectron. 2017, 87, 708–723. [Google Scholar] [CrossRef] [PubMed]
- Markom, M.A.; Shakaff, A.Y.M.; Adom, A.H.; Ahmad, M.N.; Hidayat, W.; Abdullah, A.H.; Fikri, N.A. Intelligent electronic nose system for basal stem rot disease detection. Comput. Electron. Agric. 2009, 66, 140–146. [Google Scholar] [CrossRef]
- Rizzolo, A.; Bianchi, G.; Lucido, P.; Cangelosi, B.; Pozzi, L.; Villa, G.; Clematis, F.; Pasini, C.; Curir, P. Electronic nose for the early detection of red palm weevil (Rhynchophorus ferrugineous Olivier) infestation in palms: Preliminary results. Acta Hortic. 2015, 1099, 347–355. [Google Scholar] [CrossRef]
- Fang, Y.; Ramasamy, R.P. Current and prospective methods for plant disease detection. Biosensors 2015, 4, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Mahlein, A.-K. Plant disease detection by imaging sensors—Parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef]
- Ghaffari, R.; Laothawornkitkul, J.; Zhang, F.; Iliescu, D.; Hines, E.; Leeson, M.; Napier, R.; Moore, J.P.; Paul, N.D.; Hewitt, C.N.; et al. Plant pest and disease diagnosis: Electronic nose and support vector machine approach. J. Plant Dis. Protect. 2012, 119, 200–207. [Google Scholar] [CrossRef]
- Li, G.; Fu, J.; Zhang, J.; Zheng, J. Progress in bionic information processing techniques for an electronic nose based on olfactory models. Chin. Sci. Bull. 2009, 54, 521–534. [Google Scholar] [CrossRef]
- Dudareva, N.; Negre, F.; Nagegowda, D.A.; Orlova, I. Plant volatiles: Recent advances and future perspectives. Crit. Rev. Plant Sci. 2006, 25, 417–440. [Google Scholar] [CrossRef]
- De Lacy Costello, B.P.J.; Evans, P.; Ewen, R.J.; Gunson, H.E.; Jones, P.R.H.; Ratcliffe, N.M.; Spencer-Phillips, P.T.N. Gas chromatography-mass spectrometry analyses of volatile organic compounds from potato tubers inoculated with Phytophthora infestans or Fusarium coeruleum. Plant Pathol. 2001, 50, 489–496. [Google Scholar] [CrossRef]
- Kushalappa, A.C.; Lui, L.H.; Chen, C.R.; Lee, B. Volatile fingerprinting (SPME-GC FID) to detect and discriminate diseases of potato tubers. Plant Dis. 2002, 86, 131–137. [Google Scholar] [CrossRef]
- Blasioli, S.; Biondi, E.; Samudrala, D.; Spinelli, F.; Cellini, A.; Bertaccini, A.; Cristescu, S.M.; Braschi, I. Identification of volatile markers in potato brown rot and ring rot by combined GC-MS and PTR-MS techniques: Study on in vitro and in vivo samples. J. Agric. Food Chem. 2014, 62, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Prithiviraj, B.; Hamzehzarghani, H.; Kushalappa, A.C. Volatile metabolite profiling to discriminate diseases of McIntosh apple inoculated with fungal pathogens. J. Sci. Food Agric. 2004, 84, 1333–1340. [Google Scholar] [CrossRef]
- Prithiviraj, B.; Vikram, A.; Kushalappa, A.C.; Yaylayan, V. Volatile metabolite profiling for the discrimination of onion bulbs infected by Erwinia carotovora ssp. carotovora, Fusarium oxysporum and Botrytis allii. Eur. J. Plant Pathol. 2004, 110, 371–377. [Google Scholar] [CrossRef]
- Spinelli, F.; Noferini, M.; Costa, G. Near infrared spectroscopy (NIRS): Perspective of fire blight detection in asymptomatic plant material. Acta Hortic. 2006, 704, 87–91. [Google Scholar] [CrossRef]
- Spinelli, F.; Costa, G.; Rondelli, E.; Vanneste, J.L.; Rodriguez Estrada, M.T.; Busi, S.; Savioli, S.; Cristescu, S. Volatile compounds produced by Erwinia amylovora and their potential exploitation for bacterial identification. Acta Hortic. 2011, 896, 77–84. [Google Scholar] [CrossRef]
- Cellini, A.; Biondi, E.; Buriani, G.; Farneti, B.; Rodriguez-Estrada, M.T.; Braschi, I.; Savioli, S.; Blasioli, S.; Rocchi, L.; Biasioli, F.; et al. Characterization of volatile organic compounds emitted by kiwifruit plants infected with Pseudomonas syringae pv. actinidiae and their effects on host defenses. Trees 2016, 30, 795–806. [Google Scholar] [CrossRef]
- Baietto, M.; Wilson, A.D.; Bassi, D.; Ferrini, F. Evaluation of three electronic noses for detecting incipient wood decay. Sensors 2010, 10, 1062–1092. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene role in plant growth, development and senescence: Interaction with other phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Von Dahl, C.C.; Baldwin, I.T. Deciphering the role of ethylene in plant–herbivore interactions. J. Plant Growth Regul. 2007, 26, 201–209. [Google Scholar] [CrossRef]
- Niinemets, U.; Loreto, F.; Reichstein, M. Physiological and physicochemical controls on foliar volatile organic compound emissions. Trends Plant Sci. 2004, 9, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Baldwin, E.A.; Fortuny, R.C.S.; Mattheis, J.P.; Stanley, R.; Perera, C.; Brecht, J.K. Effect of pretreatment of intact ‘Gala’ apple with ethanol vapor, hear, or 1-methylcyclopropene on quality and shelf life of fresh-cut slices. J. Am. Soc. Hortic. Sci. 2004, 129, 583–593. [Google Scholar]
- Olsson, J.; Borjesson, T.; Lundstedt, T.; Schnurer, J. Detection and quantification of ochratoxin A and deoxynivalenol in barley grains by GC–MS and electronic nose. Int. J. Food. Microbiol. 2002, 72, 203–214. [Google Scholar] [CrossRef]
- Wilson, A.D. Detection and diagnosis of bacterial wetwood in Tilia americana and Ulmus americana sapwood using a CP electronic-nose. In Industrial, Medical and Environmental Applications of Microorganisms: Current Status and Trends; Méndez-Vilas, A., Ed.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2014; pp. 209–214. ISBN 978-90-8686-243-6. [Google Scholar]
- Wilson, A.D. Bacterial wetwood detection in Fagus grandifolia and Prunus serotina sapwood using a conducting polymer electronic-nose device. In Proceedings of the V International Conference on Sensor Device Technologies and Applications, Lisbon, Portugal, 16–20 November 2014; International Academy, Research, and Industry Association (IARIA): Wilmington, DE, USA, 2014; pp. 109–113. [Google Scholar]
- Li, C.; Krewer, G.W.; Ji, P.; Scherm, H.; Kays, S.J. Gas sensor array for blueberry fruit disease detection and classification. Postharvest Biol. Technol. 2010, 55, 144–149. [Google Scholar] [CrossRef]
- Simon, J.E.; Hetzroni, A.; Bordelon, B.; Miles, G.E.; Charles, D.J. Electronic sensing of aromatic volatiles for quality sorting of blueberries. J. Food Sci. 1996, 61, 967–972. [Google Scholar] [CrossRef]
- Cheli, F.; Campagnoli, A.; Pinotti, L.; Savoini, G.; Dell’Orto, V. Electronic nose for determination of aflatoxins in maize. Biotechnol. Agron. Soc. Environ. 2009, 13, 39–43. [Google Scholar]
- Campagnoli, A.; Cheli, F.; Savoini, G.; Crotti, A.; Pastori, A.G.M.; Dell’Orto, V. Application of an electronic nose to detection of aflatoxins in corn. Vet. Res. Commun. 2009, 33, S273–S275. [Google Scholar] [CrossRef] [PubMed]
- Falasconi, M.; Gobbi, E.; Pardo, M.; Della Torre, M.; Bresciani, A.; Sberveglieri, G. Detection of toxigenic strains of Fusarium verticillioides in corn by electronic olfactory system. Sens. Actuators B 2005, 108, 250–257. [Google Scholar] [CrossRef]
- Henderson, W.G.; Khalilian, A.; Han, Y.J.; Greene, J.K.; Degenhardt, D.C. Detecting stink bugs/damage in cotton utilizing a portable electronic nose. Comput. Electron. Agric. 2010, 70, 157–162. [Google Scholar] [CrossRef]
- Blasioli, S.; Biondi, E.; Braschi, I.; Mazzucchi, U.; Bazzi, C.; Gessa, C.E. Electronic nose as an innovative tool for the diagnosis of grapevine crown gall. Anal. Chim. Acta 2010, 672, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Biondi, E.; Blasioli, S.; Fantini, M.; Braschi, I.; Bertaccini, A. Grapevine crown gall detection by electronic nose. In Proceedings of the 16th International Symposium on Olfaction and Electronic Noses, Dijon, France, 28 June–1 July 2015. [Google Scholar]
- Spinelli, F.; Noferini, M.; Vanneste, J.L.; Costa, G. Potential of the electronic-nose for the diagnosis of bacterial and fungal diseases in fruit trees. Bull. OEPP/EPPO Bull. 2010, 40, 59–67. [Google Scholar] [CrossRef]
- Wilson, A.D.; Lester, D.G. Application of aromascan analysis to detect and diagnose oak wilt in live oaks. Phytopathology 1998, 88, S97. [Google Scholar]
- Baietto, M.; Pozzi, L.; Wilson, A.D.; Bassi, D. Evaluation of a portable MOS electronic nose to detect root rots in shade tree species. Comput. Electron. Agric. 2013, 96, 117–125. [Google Scholar] [CrossRef]
- Sinha, R.; Khot, L.R.; Schroeder, B.K. FAIMS based sensing of Burkholderia cepacia caused sour skin in onions under bulk storage condition. Food Meas. 2017, 11, 1578–1585. [Google Scholar] [CrossRef]
- Baldwin, E.; Plotto, A.; Manthey, J.; McCollum, G.; Bai, J.; Irey, M.; Cameron, R.; Luzio, G. Effect of Liberibacter infection (Huanglongbing disease) of citrus on orange fruit physiology and fruit/fruit juice quality: Chemical and physical analyses. J. Agric. Food Chem. 2010, 58, 1247–1262. [Google Scholar] [CrossRef] [PubMed]
- Pallottino, F.; Costa, C.; Antonucci, F.; Strano, M.C.; Calandra, M.; Solaini, S.; Menesatti, P. Electronic nose application for determination of Penicillium digitatum in Valencia oranges. J. Sci. Food Agric. 2012, 92, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.; Hepher, M.J.; Sommerville, J. Detection of Serpula lacrymans infestation with a polypyrrole sensor array. Sens. Actuator B Chem. 2006, 113, 989–997. [Google Scholar] [CrossRef]
- Stinson, J.A.; Persaud, K.C.; Bryning, G. Generic system for the detection of statutory potato pathogens. Sens. Actuators B Chem. 2006, 116, 100–106. [Google Scholar] [CrossRef]
- Biondi, E.; Blasioli, S.; Galeone, A.; Spinelli, F.; Cellini, A.; Lucchese, C.; Braschi, I. Detection of potato brown rot and ring rot by electronic nose: From laboratory to real scale. Talanta 2014, 129, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Rutolo, M.F.; Clarkson, J.P.; Covington, J.A. The use of an electronic nose to detect early signs of soft-rot infection in potatoes. Biosyst. Eng. 2018, 167, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Wang, J. Use of electronic nose technology for identifying rice infestation by Nilaparvata lugens. Sens. Actuators B Chem. 2011, 160, 15–21. [Google Scholar] [CrossRef]
- Xu, S.; Zhou, Z.; Lu, H.; Luo, X.; Lan, Y.; Zhang, Y.; Li, Y. Estimation of the age and amount of brown rice plant hoppers based on bionic electronic nose use. Sensors 2014, 14, 18114–18130. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhang, W.; Zhu, N.; Mao, S.; Tu, K. Early detection and classification of pathogenic fungal disease in post-harvest strawberry fruit by electronic nose and gas chromatography–mass spectrometry. Food Res. Int. 2014, 62, 162–168. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Cheng, S. Predicting attacked time of tomato seedling by e-nose based on kernel principal component analysis. In Proceedings of the ASABE Annual International Meeting, Orlando, FL, USA, 17–20 July 2016. [Google Scholar]
- Paolesse, R.; Alimelli, A.; Martinelli, E.; Di Natale, C.; D’Amico, A.; D’Egidio, M.G.; Aureli, G.; Ricelli, A.; Fanelli, C. Detection of fungal contamination of cereal grain samples by an electronic nose. Sens. Actuators B Chem. 2006, 119, 425–430. [Google Scholar] [CrossRef]
- Eifler, J.; Martinelli, E.; Santonico, M.; Capuano, R.; Schild, D.; Di Natale, C. Differential detection of potentially hazardous Fusarium species in wheat grains by an electronic nose. PLoS ONE 2011, 6, e21026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, J. Detection of age and insect damage incurred by wheat, with an electronic nose. J. Stored Prod. Res. 2007, 43, 489–495. [Google Scholar] [CrossRef]
- Naznin, H.A.; Kiyohara, D.; Kimura, M.; Miyazawa, M.; Shimizu, M.; Hyakumachi, M. Systemic resistance induced by volatile organic compounds emitted by plant growth-promoting fungi in Arabidopsis thaliana. PLoS ONE 2014, 9, e86882. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.M.; Wildt, J.; Kappers, I.F.; Bouwmeester, H.J.; Hofstee, J.W.; van Henten, E.J. Detection of diseased plants by analysis of volatile organic compound emission. Annu. Rev. Phytopathol. 2011, 49, 157–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanchiswamy, C.N.; Malnoy, M.; Maffei, M.E. Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 2015, 6, 151. [Google Scholar] [CrossRef] [PubMed]
- Bitas, V.; Kim, H.; Bennett, J.W.; Kang, S. Sniffing on microbes: Diverse roles of microbial volatile organic compounds in plant health. Mol. Plant-Microbe Interact. 2013, 26, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Angle, C.; Waggoner, L.P.; Ferrando, A.; Haney, P.; Passler, T. Canine detection of the volatilome: A review of implications for pathogen and disease detection. Front. Vet. Sci. 2016, 3, 47. [Google Scholar] [CrossRef] [PubMed]
- Zuckerman, D.; Yttri, J. Antibiotics: When Science and Wishful Thinking Collide. Health Affairs Blogg. Available online: http://www.center4research.org/antibiotics-science-wishful-thinking-collide (accessed on 21 August 2018).
- Shivaprasad, H.L.; Palmieri, C. Pathology of mycobacteriosis in birds. The Veterinary Clinics of North America. Exot. Anim. Pract. 2012, 15, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Reavill, D.R.; Schmidt, R.E. Mycobacterial lesions in fish, amphibians, reptiles, rodents, lagomorphs, and ferrets with reference to animal models. The Veterinary Clinics of North America. Exot. Anim. Pract. 2012, 15, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Wobeser, G.A. Essentials of Disease in Wild Animals, 1st ed.; Blackwell Publishing: Ames, IA, USA, 2006; p. 170. ISBN 978-0-8138-0589-4. [Google Scholar]
- Ryan, T.J.; Livingstone, P.G.; Ramsey, D.S.; de Lisle, G.W.; Nugent, G.; Collins, D.M.; Buddle, B.M. Advances in understanding disease epidemiology and implications for control and eradication of tuberculosis in livestock: The experience from New Zealand. Vet. Microbiol. 2006, 112, 211–219. [Google Scholar] [CrossRef] [PubMed]
- White, P.C.; Böhm, M.; Marion, G.; Hutchings, M.R. Control of bovine tuberculosis in British livestock: There is no ‘silver bullet’. Trends Microbiol. 2008, 16, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.I.; Judge, J.; Delahay, R.J. Farm husbandry and badger behaviour: Opportunities to manage badger to cattle transmission of Mycobacterium bovis? Prev. Vet. Med. 2010, 93, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Holt, N. The Infected Elephant in the Room. Slate’s Animal Blog. Available online: http://www.slate.com/blogs/wild_things/2015/03/24/elephant_tuberculosis_epidemic_zoo_and_circus_animals_passing_tb_to_humans.html (accessed on 24 August 2018).
- Schwarz, C.M. The Chambers Dictionary; Allied Chambers India Ltd.: Edinburg, UK, 1998; p. 352. ISBN 978-81-86062-25-8. [Google Scholar]
- Mandell, G.; Bennett, J.; Dolin, R. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 7th ed.; Churchill Livingstone; Elsevier: Philadelphia, PA, USA, 2010; Chapter 250; ISBN 978-0-443-06839-3. [Google Scholar]
- WHO Model Formulary. Available online: http://apps.who.int/medicinedocs/documents/s16879e/s16879e.pdf (accessed on 21 August 2018).
- Fend, R.; Geddes, R.; Lesellier, S.; Vordermeier, H.-M.; Corner, L.A.L.; Gormley, E.; Costello, E.; Hewinson, R.G.; Marlin, D.J.; Woodman, A.C.; et al. Use of an electronic nose to diagnose Mycobacterium bovis infection in badgers and cattle. J. Clin. Microbiol. 2005, 43, 1745–1751. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.D.; Forse, L.B. Discrimination between Pseudogymnoascus destructans, other dermatophytes of cave-dwelling bats, and related innocuous keratinophilic fungi based on electronic-nose/GC signatures of VOC-metabolites produced in culture. In Proceedings of the VIII International Conference on Sensor Device Technologies and Applications, Rome, Italy, 10–14 September 2017; International Academy, Research, and Industry Association (IARIA): Wilmington, DE, USA, 2017; pp. 5–11. [Google Scholar]
- Wilson, A.D.; Forse, L.B. Differences in VOC-metabolite profiles of Pseudogymnoascus destructans and related fungi by electronic-nose/GC analyses of headspace volatiles derived from axenic cultures. Sens. Transducers 2018, 220, 9–19. [Google Scholar]
- Wilson, A.D.; Oberle, C.S.; Oberle, D.F. Detection of off-flavor in catfish using a conducting polymer electronic-nose technology. Sensors 2013, 13, 15968–15984. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.B.; Wang, S.Z.; Yin, Y.G.; Hoffmann, W.C.; Zheng, X.Z. Using a surface plasmon resonance biosensor for rapid detection of Salmonella typhimurium in chicken carcass. J. Bionic Eng. 2008, 5, 239–246. [Google Scholar] [CrossRef]
- Knobloch, H.; Schroedl, W.; Turner, C.; Chambers, M.; Reinhold, P. Electronic nose responses and acute phase proteins correlate in blood using a bovine model of respiratory infection. Sens. Actuators B 2010, 144, 81–87. [Google Scholar] [CrossRef]
- Mottram, T.T.; Dobbelaar, P.; Schukken, Y.H.; Hobbs, P.J.; Bartlett, P.N. An experiment to determine the feasibility of automatically detecting hyperketonaemia in dairy cows. Livest. Prod. Sci. 1999, 61, 7–11. [Google Scholar] [CrossRef]
- Wlodzimirow, K.A.; Abu-Hanna, A.; Schultz, M.J.; Maas, M.A.; Bos, L.D.; Sterk, P.J.; Knobel, H.H.; Soers, R.J.; Chamuleau, R.A. Exhaled breath analysis with electronic nose technology for detection of acute liver failure in rats. Biosens. Bioelectron. 2014, 53, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Cramp, A.P.; Sohn, J.H.; James, P.J. Detection of cutaneous myiasis in sheep using an ‘electronic nose’. Vet. Parasitol. 2009, 166, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Van Soolingen, D.; Hoogenboezem, T.; de Haas, P.E.; Hermans, P.W.; Koedam, M.A.; Teppema, K.S.; Brennan, P.J.; Besra, G.S.; Portaels, F.; Top, J.; et al. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: Characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 1997, 47, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Niemann, S.; Rüsch-Gerdes, S.; Joloba, M.L.; Whalen, C.C.; Guwatudde, D.; Ellner, J.J.; Eisenach, K.; Fumokong, N.; Johnson, J.L.; Aisu, T.; et al. Mycobacterium africanum subtype II is associated with two distinct genotypes and is a major cause of human tuberculosis in Kampala, Uganda. J. Clin. Microbiol. 2002, 40, 3398–3405. [Google Scholar] [CrossRef] [PubMed]
- Niobe-Eyangoh, S.N.; Kuaban, C.; Sorlin, P.; Cunin, P.; Thonnon, J.; Sola, C.; Rastogi, N.; Vincent, V.; Gutierrez, M.C. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 2003, 41, 2547–2553. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.A. Mycobacterial infections in reptiles. Vet. Clin. Exot. Anim. 2012, 15, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Abbas, A.K.; Fausto, N.; Mitchell, R.N. Robbins Basic Pathology, 8th ed.; Saunders; Elsevier: Philadelphia, PA, USA, 2007; pp. 516–522. ISBN 978-1-4160-2973-1. [Google Scholar]
- Thoen, C.; Lobue, P.; de Kantor, I. The importance of Mycobacterium bovis as a zoonosis. Vet. Microbiol. 2006, 112, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Acton, Q.A. Mycobacterium Infections: New Insights for the Healthcare Professional; ScholarlyEditions: Atlanta, GA, USA, 2011; p. 1968. ISBN 978-1-4649-0122-5. [Google Scholar]
- Pfyffer, G.E.; Auckenthaler, R.; van Embden, J.D.; van Soolingen, D. Mycobacterium canettii, the smooth variant of M. tuberculosis, isolated from a Swiss patient exposed in Africa. Emerg. Infect. Dis. 1998, 4, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Panteix, G.; Gutierrez, M.C.; Boschiroli, M.L.; Rouviere, M.; Plaidy, A.; Pressac, D.; Porcheret, H.; Chyderiotis, G.; Ponsada, M.; Van Oortegem, K.; et al. Pulmonary tuberculosis due to Mycobacterium microti: A study of six recent cases in France. J. Med. Microbiol. 2010, 59, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. American Thoracic Society, Medical Section of the American Lung Association. Am. J. Respir. Crit. Care Med. 1997, 156, S1–S25. [Google Scholar] [CrossRef] [PubMed]
- Bento, J.; Silva, A.S.; Rodrigues, F.; Duarte, R. Diagnostic tools in tuberculosis. Acta Med. Port. 2011, 24, 145–154. [Google Scholar] [PubMed]
- Escalante, P. In the clinic. Tuberculosis. Ann. Intern. Med. 2009, 150, ITC61-614, quiz ITV616. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, R.; Wang, J.; Liu, L.; Zheng, Y.; Shen, Y.; Qi, T.; Lu, H. Interferon-gamma release assays for the diagnosis of active tuberculosis in HIV-infected patients: A systematic review and meta-analysis. PLoS ONE 2011, 6, e26827. [Google Scholar] [CrossRef] [PubMed]
- Foundation for Innovative New Diagnostics. Diagnostics for Tuberculosis: Global Demand and Market Potential; Special Programme for Research and Training in Tropical Diseases; World Health Organization: Geneva, Switzerland, 2006; pp. 36–37. ISBN 978-92-4-156330-7. [Google Scholar]
- National Collaborating Centre for Chronic Conditions (UK). Tuberculosis: Clinical Diagnosis and Management of Tuberculosis, and Measures for Its Prevention and Control; TB Clinical Guideline 117; National Institute for Health and Clinical Excellence: London, UK, 2011; pp. 42–121. ISBN 978-1-84936-537-6. [Google Scholar]
- Steingart, K.R.; Flores, L.L.; Dendukuri, N.; Schiller, I.; Laal, S.; Ramsay, A.; Hopewell, P.C.; Pai, M. Commercial serological tests for the diagnosis of active pulmonary and extrapulmonary tuberculosis: An updated systematic review and meta-analysis. PLoS Med. 2011, 8, e1001062. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, J.Z.; Everett, C.K.; Steingart, K.R.; Cattamanchi, A.; Huang, L.; Hopewell, P.C.; Pai, M. Interferon-γ release assays for active pulmonary tuberculosis diagnosis in adults in low- and middle-income countries: Systematic review and meta-analysis. J. Infect. Dis. 2011, 204 (Suppl. 4), S1120–S1129. [Google Scholar] [CrossRef] [PubMed]
- Sester, M.; Sotgiu, G.; Lange, C.; Giehl, C.; Girardi, E.; Migliori, G.B.; Bossink, A.; Dheda, K.; Diel, R.; Dominguez, J.; et al. Interferon-γ release assays for the diagnosis of active tuberculosis: A systematic review and meta-analysis. Eur. Respir. J. 2011, 37, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, H.; Köhler, N.; Reinhold, P.; Turner, C.; Chambers, M. Volatile organic compound (VOC) analysis for disease detection: Proof of principle for field studies detecting paratuberculosis and brucellosis. AIP Conf. Proc. 2009, 1137, 195–197. [Google Scholar] [CrossRef]
- Phillips, M.; Cataneo, R.N.; Condos, R.; Ring Erickson, G.A.; Greenberg, J.; La Bombardi, V.; Munawar, M.I.; Tietje, O. Volatile biomarkers of pulmonary tuberculosis in the breath. Tuberculosis 2007, 87, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.; Basa-Dalay, V.; Bothamley, G.; Cataneo, R.N.; Lam, P.K.; Natividad, M.P.; Schmitt, P.; Wai, J. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis 2010, 90, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Du Preez, I.; Loots, D.T. New sputum metabolite markers implicating adaptations of the host to Mycobacterium tuberculosis, and vice versa. Tuberculosis 2013, 93, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Bruins, M.; Rahim, Z.; Bos, A.; van de Sande, W.W.J.; Endtz, H.P.; van Belkum, A. Diagnosis of active tuberculosis by e-nose analysis of exhaled air. Tuberculosis 2013, 93, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Gerber, N.N.; Lechevalier, H.A. Geosmin, an earthy-smelling compound isolated from actinomycetes. Appl. Microbiol. 1965, 13, 935–938. [Google Scholar] [PubMed]
- Grimm, C.C.; Lloyd, S.W.; Batista, R.; Zimba, P.V. Using microwave distillation-solid-phase-microextraction-gas chromatography-mass spectrometry for analyzing fish tissue. J. Chromatogr. Sci. 2000, 38, 289–296. [Google Scholar] [CrossRef] [PubMed]
- de Heer, K.; Kok, M.G.M.; Fens, N.; Weersink, E.J.M.; Zwinderman, A.H.; van der Schee, M.P.C.; Visser, C.E.; van Oers, M.H.J.; Sterk, P.J. Detection of airway colonization by Aspergillus fumigatus by use of electronic nose technology in patients with cystic fibrosis. J. Clin. Microbiol. 2016, 54, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Dettenkofer, M.; Wenzler-Röttele, S.; Babikir, R.; Bertz, H.; Ebner, W.; Meyer, E.; Rüden, H.; Gastmeier, P.; Daschner, F.D. Surveillance of nosocomial sepsis and pneumonia in patients with a bone marrow or peripheral blood stem cell transplant: A multicenter project. Clin. Infect. Dis. 2005, 40, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Cornillet, A.; Camus, C.; Nimubona, S.; Gandemer, V.; Tattevin, P.; Belleguic, C.; Chevrier, S.; Meunier, C.; Lebert, C.; Aupe, M.; et al. Comparison of epidemiological, clinical, and biological features of invasive aspergillosis in neutropenic and nonneutropenic patients: A 6-year survey. Clin. Infect. Dis. 2006, 43, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Robenshtok, E.; Gafter-Gvili, A.; Goldberg, E.; Weinberger, M.; Yeshurun, M.; Leibovici, L.; Paul, M. Antifungal prophylaxis in cancer patients after chemotherapy or hematopoietic stem-cell transplantation: Systematic review and meta-analysis. J. Clin. Oncol. 2007, 25, 5471–5489. [Google Scholar] [CrossRef] [PubMed]
- Barth, P.J.; Rossberg, C.; Koch, S.; Ramaswamy, A. Pulmonary aspergillosis in an unselected autopsy series. Pathol. Res. Pract. 2000, 196, 73–80. [Google Scholar] [CrossRef]
- Groll, A.H.; Shah, P.M.; Mentzel, C.; Schneider, M.; Just-Nuebling, G.; Huebner, K. Trends in the postmortem epidemiology of invasive fungal infections at a university hospital. J. Infect. 1996, 33, 23–32. [Google Scholar] [CrossRef]
- Aisner, J.; Wiernik, P.H.; Schimpff, S.C. Treatment of invasive aspergillosis: Relation of early diagnosis and treatment to response. Ann. Intern. Med. 1977, 86, 539–543. [Google Scholar] [CrossRef] [PubMed]
- von Eiff, M.; Roos, N.; Schulten, R.; Hesse, M.; Zuhlsdorf, M.; van de Loo, J. Pulmonary aspergillosis: Early diagnosis improves survival. Respiration 1995, 62, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.L.; Chen, Y.Q.; Wang, K.; Qin, S.M.; Wu, C.; Kong, J.L. Accuracy of BAL galactomannan in diagnosing invasive aspergillosis: A bivariate metaanalysis and systematic review. Chest 2010, 138, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Tukey, M.H.; Wiener, R.S. Population-based estimates of transbronchial lung biopsy utilization and complications. Respir. Med. 2012, 106, 1559–1565. [Google Scholar] [CrossRef] [PubMed]
- Carr, I.M.; Koegelenberg, C.F.N.; von Groote-Bidlingmaier, F.; Mowlana, A.; Silos, K.; Haverman, T.; Diacon, A.H.; Bolliger, C.T. Blood loss during flexible bronchoscopy: A prospective observational study. Respiration 2012, 84, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Facciolongo, N.; Patelli, M.; Gasparini, S.; Lazzari, A.L.; Salio, M.; Simonassi, C.; Del Prato, B.; Zanoni, P. Incidence of complications in bronchoscopy: Multicentre prospective study of 20,986 bronchoscopies. Monaldi Arch. Chest Dis. 2009, 71, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Mu, D.; Chu, D.; Fu, E.; Xie, Y.; Liu, T. Severe complications of bronchoscopy. Respiration 2008, 76, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Pue, C.A.; Pacht, E.R. Complications of fiberoptic bronchoscopy at a university hospital. Chest 1995, 107, 430–432. [Google Scholar] [CrossRef] [PubMed]
- Sibila, O.; Garcia-Bellmunt, L.; Giner, J.; Merino, J.L.; Suarez-Cuartin, G.; Torrego, A.; Solanes, I.; Castillo, D.; Valera, J.L.; Cosio, B.G.; et al. Identification of airway bacterial colonization by an electronic nose in chronic obstructive pulmonary disease. Respir. Med. 2014, 108, 1608–1614. [Google Scholar] [CrossRef] [PubMed]
- Shafiek, H.; Fiorentino, F.; Merino, J.L.; López, C.; Oliver, A.; Segura, J.; de Paul, I.; Sibila, O.; Agustí, A.; Cosío, B.G. Using the electronic nose to identify airway infection during COPD exacerbations. PLoS ONE 2015, 10, e0135199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragonieri, S.; Quaranta, V.N.; Carratu, P.; Ranieri, T.; Marra, L.; D’Alba, G.; Resta, O. An electronic nose may sniff out amyotrophic lateral sclerosis. Resp. Physiol. Neurobiol. 2016, 232, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.A.; Thomas, P.S.; Stone, E.; Lewis, C.; Yates, D.H. A breath test for malignant mesothelioma using an electronic nose. Eur. Respir. J. 2012, 40, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Brekelmans, M.P.; Fens, N.; Brinkman, P.; Bos, L.D.; Sterk, P.J.; Tak, P.P.; Gerlag, D.M. Smelling the Diagnosis: The Electronic Nose as Diagnostic Tool in Inflammatory Arthritis. A Case-Reference Study. PLoS ONE 2016, 11, e0151715. [Google Scholar] [CrossRef] [PubMed]
- Dragonieri, S.; Schot, R.; Mertens, B.J.; Le Cessie, S.; Gauw, S.A.; Spanevello, A.; Resta, O.; Willard, N.P.; Vink, T.J.; Rabe, K.F.; et al. An electronic nose in the discrimination of patients with asthma and controls. J. Allergy Clin. Immunol. 2007, 120, 856–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montuschi, P.; Santonico, M.; Mondino, C.; Pennazza, G.; Mantini, G.; Martinelli, E.; Capuano, R.; Ciabattoni, G.; Paolesse, R.; Di Natale, C.; et al. Diagnose performance of an electronic nose, fractional exhaled nitric oxide and lung function testing in asthma. Chest 2010, 137, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Aathithan, S.; Plant, J.C.; Chaudry, A.N.; French, G.L. Diagnosis of bacteriuria by detection of volatile organic compounds in urine using an automated headspace analyzer with multiple conducting polymer sensors. J. Clin. Microbiol. 2001, 39, 2590–2593. [Google Scholar] [CrossRef] [PubMed]
- Covington, J.A.; Westenbrink, E.W.; Ouaret, N.; Harbord, R.; Bailey, C.; O’Connell, N.; Cullis, J.; Williams, N.; Nwokolo, C.U.; Bardhan, K.D.; et al. Application of a novel tool for diagnosing bile acid diarrhoea. Sensors 2013, 13, 11899–11912. [Google Scholar] [CrossRef] [PubMed]
- Hay, P.; Tummon, A.; Ogunfile, M.; Adebiyi, A.; Adefowora, A. Evaluation of a novel diagnostic test for bacterial vaginosis: The electronic nose. Int. J. STD AIDS 2003, 14, 114–118. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, A.; Pennazza, G.; Santonico, M.; Martinelli, E.; Roscioni, C.; Galluccio, G.; Paolesse, R.; Di Natale, C. An investigation on electronic nose diagnosis of lung cancer. Lung Cancer 2010, 68, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Joensen, O.; Paff, T.; Haarman, E.G.; Skovgaard, I.M.; Jensen, P.Ø.; Bjarnsholt, T.; Nielsen, K.G. Exhaled breath analysis using electronic nose in cystic fibrosis and primary ciliary dyskinesia patients with chronic pulmonary infections. PLoS ONE 2014, 9, e115584. [Google Scholar] [CrossRef] [PubMed]
- Saidi, T.; Zaima, O.; Moufida, M.; El Barib, N.; Ionescuc, R.; Bouchikhi, B. Exhaled breath analysis using electronic nose and gas chromatography–mass spectrometry for non-invasive diagnosis of chronic kidney disease, diabetes mellitus and healthy subjects. Sens. Actuators B 2018, 257, 178–188. [Google Scholar] [CrossRef]
- Westenbrink, E.; Arasaradnam, R.P.; O’Connell, N.O.; Bailey, C.; Nwokolo, C.; Bardhan, K.D.; Covington, J.A. Development and application of a new electronic nose instrument for the detection of colorectal cancer. Biosens. Bioelectron. 2015, 67, 733–738. [Google Scholar] [CrossRef] [PubMed]
- De Meij, T.G.; Larbi, I.B.; van der Schee, M.P.; Lentferink, Y.E.; Paff, T.; Terhaar Sive Droste, J.S.; Mulder, C.J.; van Bodegraven, A.A.; de Boer, N.K. Electronic nose can discriminate colorectal carcinoma and advanced adenomas by fecal volatile biomarker analysis: Proof of principle study. Int. J. Cancer 2014, 134, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Hakim, M.; Broza, Y.Y.; Billan, S.; Abdah-Bortnyak, R.; Kuten, A.; Tisch, U.; Haick, H. Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br. J. Cancer 2010, 103, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, N.; Zakaria, A.; Omar, M.I.; Shakaff, A.Y.; Masnan, M.J.; Kamarudin, L.M.; Rahim, N.A.; Zakaria, N.Z.I.; Abdullah, A.A.; Othman, A.; et al. In-vitro diagnosis of single and poly microbial species targeted for diabetic foot infection using e-nose technology. BMC Bioinform. 2015, 16, 158. [Google Scholar] [CrossRef] [PubMed]
- Boilot, P.; Hines, E.; Gardner, J.; Pitt, R.; John, S.; Mitchell, J.; Morgan, D.W. Classification of bacteria responsible for ENT and eye infections using the Cyranose system. IEEE Sens. J. 2002, 2, 247–253. [Google Scholar] [CrossRef]
- Hakim, M.; Billan, S.; Tisch, U.; Peng, G.; Dvrokind, I.; Marom, O.; Abdah-Bortnya, R.; Kuten, A.; Haick, H. Diagnosis of head-and-neck cancer from exhaled breath. Br. J. Cancer 2011, 104, 1649–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Covington, J.; Harbord, R.; Westenbrink, E.W.; Bailey, C.; O’Connell, N.; Dhaliwal, A.; Nwokolo, C.; Foley, A.; Marya, N.B.; Baptista, V.; et al. Detection of urinary volatile organic compounds in patients with inflammatory bowel disease and controls by an electronic nose—A transatlantic study. Gastroenterology 2014, 146, S795–S796. [Google Scholar] [CrossRef]
- De Meij, T.G.J.; de Boer, N.K.H.; Benninga, M.A.; Lentferink, Y.E.; de Groot, E.F.J.; van de Velde, M.E.; van Bodegraven, A.A.; van der Scheea, M.P. Faecal gas analysis by electronic nose as a novel, non-invasive method for assessment of active and quiescent paediatric inflammatory bowel disease: Proof of principle study. J. Crohn’s Colitis 2014, 8, 91–106. [Google Scholar] [CrossRef]
- Shepherd, S.F.; McGuire, N.D.; de Lacy Costello, B.P.; Ewen, R.J.; Jayasena, D.H.; Vaughan, K.; Ahmed, I.; Probert, C.S.; Ratcliffe, N.M. The use of a gas chromatograph coupled to a metal oxide sensor for rapid assessment of stool samples from irritable bowel syndrome and inflammatory bowel disease patients. J. Breath Res. 2014, 8, 026001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monasta, L.; Pierobon, C.; Princivalle, A.; Martelossi, S.; Marcuzzi, A.; Pasini, F.; Perbellini, L. Inflammatory bowel disease and patterns of volatile organic compounds in the exhaled breath of children: A case-control study using Ion Molecule Reaction-Mass Spectrometry. PLoS ONE 2017, 12, e0184118. [Google Scholar] [CrossRef] [PubMed]
- McGuire, N.D.; Ewen, R.J.; Costello, C.D.; Garner, C.E.; Probert, C.S.J.; Vaughan, K.; Ratcliffe, N.M. Towards point of care testing for C. difficile infection by volatile profiling, using the combination of a short multi-capillary gas chromatography column with metal oxide detection. Meas. Sci. Technol. 2014, 25, 065108. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Anguri, H.; Nonaka, A.; Kataoka, K.; Nagata, H.; Kita, J.; Shizukuishi, S. Clinical assessment of oral malodor by the electronic nose system. J. Dent. Res. 2004, 83, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Kharitonov, S. Exhaled markers of inflammatory lung disease, ready for routine monitoring. Swiss Med. Wkly. 2004, 134, 175–192. [Google Scholar] [PubMed]
- De Heer, K.; van der Schee, M.P.; Zwinderman, K.; van den Berk, I.A.; Visser, C.E.; van Oers, R.; Sterk, P.J. Invasive pulmonary aspergillosis in prolonged chemotherapy-induced neutropenia: A proof-of-principle study. J. Clin. Microbiol. 2013, 51, 1490–1495. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Takahashi, Y.; Konishi, K.; Katsuumi, I. Association of odor from infected root canal analyzed by an electronic nose with isolated bacteria. J. Endod. 2007, 33, 1106–1109. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, D.J.C.; Niemark, H.J.; Buijck, M.; van Weissenbruch, M.M.; Brinkman, P.; Benninga, M.A.; van Kaam, A.H.; Kramer, B.W.; Andriessen, P.; de Boer, N.K.H.; et al. Detection of sepsis in preterm infants by fecal volatile organic compounds analysis: A proof of principle study. J. Pediatr. Gastroenterol. Nutr. 2017, 65, e47–e52. [Google Scholar] [CrossRef] [PubMed]
- De Gennaro, G.; Dragonieri, S.; Longobardi, F.; Musti, M.; Stallone, G.; Trizio, L.; Tutino, M. Chemical characterization of exhaled breath to differentiate between patients with malignant pleural mesothelioma from subjects with similar professional asbestos exposure. Anal. Bioanal. Chem. 2010, 398, 3043–3050. [Google Scholar] [CrossRef] [PubMed]
- De Meij, T.G.; van der Schee, M.P.; Berkhout, D.J.; van de Velde, M.E.; Jansen, A.E.; Kramer, B.W.; van Weissenbruch, M.M.; van Kaam, A.H.; Andriessen, P.; van Goudoever, J.B.; et al. Early detection of necrotizing enterocolitis by fecal volatile organic compounds analysis. J. Pediatr. 2015, 167, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Finberg, J.P.M.; Schwartz, M.; Jeries, R.; Badarny, S.; Nakhleh, M.K.; Abu Daoud, E.; Ayubkhanov, Y.; Aboud-Hawa, M.; Broza, Y.Y.; Haick, H. Sensor array for detection of early stage Parkinson’s disease before medication. ACS Chem. Neurosci. 2018, in press. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-Y.; Peng, H.-Y.; Chang, C.-J.; Chen, P.-C. Diagnostic accuracy of breath tests for pneumoconiosis using an electronic nose. J. Breath Res. 2017, 12, 016001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hockstein, N.G.; Thaler, E.R.; Torigian, D.; Miller, W.T., Jr.; Deffenderfer, O.; Hanson, C.W. Diagnosis of pneumonia with an electronic nose: Correlation of vapor signature with chest computed tomography scan findings. Laryngoscope 2004, 114, 1701–1705. [Google Scholar] [CrossRef] [PubMed]
- Hanson, C.W., III; Thaler, E.R. Electronic nose prediction of a clinical pneumonia score: Biosensors and microbes. Anesthesiology 2005, 102, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Dragonieri, S.; Brinkman, P.; Mouw, E.; Zwinderman, A.H.; Carratu´, P.; Resta, O.; Sterk, P.J.; Jonkers, R.E. An electronic nose discriminates exhaled breath of patients with untreated pulmonary sarcoidosis from controls. Respir. Med. 2013, 107, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Dutta, R.; Morgan, D.; Baker, N.; Gardner, J.W.; Hines, E.L. Identification of Staphylococcus aureus infections in hospital environment: Electronic nose based approach. Sens. Actuators B 2005, 109, 335–362. [Google Scholar] [CrossRef]
- Vaira, D.; Holton, J.; Ricci, C.; Basset, C.; Gatta, L.; Perna, F.; Tampieri, A.; Miglioli, M. Helicobacter pylori infection from pathogenesis to treatment: A critical reappraisal. Aliment. Pharm. Ther. 2002, 16, 105–113. [Google Scholar] [CrossRef]
- Lai, S.Y.; Deffenderfer, O.F.; Hanson, W.; Phillips, M.P.; Thaler, E.R. Identification of upper respiratory bacterial pathogens with the electronic nose. Laryngoscope 2002, 112, 975–979. [Google Scholar] [CrossRef] [PubMed]
- Roine, A.; Saviauk, T.; Kumpulainen, P.; Karjalainen, M.; Tuokko, A.; Aittoniemi, J.; Vuento, R.; Lekkala, J.; Lehtima, T.; Tammela, T.L.; et al. Rapid and accurate detection of urinary pathogens by mobile IMS-based electronic nose: A Proof-of-principle study. PLoS ONE 2014, 9, e114279. [Google Scholar] [CrossRef] [PubMed]
- Hockstein, N.G.; Thaler, E.R.; Lin, Y.; Lee, D.D.; Hanson, C.W. Correlation of pneumonia score with electronic nose signature: A prospective study. Ann. Otol. Rhinol. Laryngol. 2005, 114, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, R.M.; Boumans, M.L.L.; Smolinska, A.; Stobberingh, E.E.; Kaufmann, R.; Roekaerts, P.M.H.J.; Bergmans, D.C.J.J. Electronic nose analysis of exhaled breath to diagnose ventilator associated pneumonia. Respir. Med. 2015, 109, 1454–1459. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Xu, X.; Shen, Y.; Yan, J.; He, Q.; Ma, J.; Liu, T. Detection of wound pathogen by an intelligent electronic nose. Sens. Mater. 2009, 21, 155–166. [Google Scholar]
- Saviauk, T.; Kiiski, J.P.; Nieminen, M.K.; Tamminen, N.N.; Roine, A.N.; Kumpulainen, P.S.; Hokkinen, L.J.; Karjalainen, M.T.; Vuento, R.E.; Aittoniemi, J.J.; et al. Electronic nose in the detection of wound infection bacteria from bacterial cultures: A proof-of-principle study. Eur. Surg. Res. 2018, 59, 1–11. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Surveillance, Prevention and Control of Chronic Respiratory Diseases: A Comprehensive Approach. Available online: http://www.who.int/respiratory/publications/global_surveillance/en (accessed on 27 August 2018).
- Balmes, J.; Becklakem, M.; Blanc, P.; Henneberger, P.; Kreiss, K.; Mapp, C.; Milton, D.; Schwartz, D.; Toren, K.; Viegi, G. American Thoracic Society statement: Occupational contribution to the burden of airway disease. Am. J. Respir. Crit. Care Med. 2003, 167, 787–797. [Google Scholar] [CrossRef]
- Cohen, R.A. Resurgent coal mine dust lung disease: Wave of the future or a relic of the past? Occup. Environ. Med. 2016, 73, 715–716. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.K.; Gupta, R.K. Asbestos in developing countries: Magnitude of risk and its practical implications. Int. J. Occup. Med. Environ. Health 2004, 17, 179–185. [Google Scholar] [CrossRef]
- Capelli, L.; Taverna, G.; Bellini, A.; Eusebio, L.; Buffi, N.; Lazzeri, M.; Guazzoni, G.; Bozzini, G.; Seveso, M.; Mandressi, A.; et al. Application and uses of electronic noses for clinical diagnosis on urine samples: A review. Sensors 2016, 16, 1708. [Google Scholar] [CrossRef] [PubMed]
- Horstmann, M.; Steinbach, D.; Fischer, C.; Enkelmann, A.; Grimm, M.; Voss, A. An electronic nose system detects bladder cancer in urine specimen: First results of a pilot study. J. Urol. 2015, 193, 560–561. [Google Scholar] [CrossRef]
- Pendharkar, S.; Mehta, S. The clinical significance of exhaled nitric oxide in asthma. Can. Respir. J. 2008, 15, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Prado, C.M.; Martins, M.A.; Tibério, I.F. Nitric oxide in asthma physiopathology. ISRN Allergy 2011, 19, 832560. [Google Scholar] [CrossRef] [PubMed]
- Lacey, J.N.; Kidel, C.; van der Kaaij, J.M.; Brinkman, P.; Gilbert-Kawai, E.T.; Grocott, M.P.W.; Mythen, M.G.; Martin, D.S. The Smell of Hypoxia: Using an electronic nose at altitude and proof of concept of its role in the prediction and diagnosis of acute mountain sickness. Physiol. Rep. 2018, 6, e13854. [Google Scholar] [CrossRef] [PubMed]
- Lüscher, T.F.; Barton, M. Biology of the endothelium. Clin. Cardiol. 1997, 20, 3–10. [Google Scholar]
- Ma, F.X.; Zhou, B.; Chen, Z.; Ren, Q.; Lu, S.H.; Sawamura, T.; Han, Z.C. Oxidized low density lipoprotein impairs endothelial progenitor cells by regulation of endothelial nitric oxide synthase. J. Lipid Res. 2006, 47, 1227–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abramson, S.B. Nitric oxide in inflammation and pain associated with osteoarthritis. Arthritis Res. Ther. 2008, 10, S2. [Google Scholar] [CrossRef] [PubMed]
- Maki-Petaja, K.M.; Cheriyan, J.; Booth, A.D.; Hall, F.C.; Brown, J.; Wallace, S.M.; Ashby, M.J.; McEniery, C.M.; Wilkinson, I.B. Inducible nitric oxide synthase activity is increased in patients with rheumatoid arthritis and contributes to endothelial dysfunction. Int. J. Cardiol. 2008, 129, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Van der Vliet, A.; Eiserich, J.P.; Cross, C.E. Nitric oxide: A pro-inflammatory mediator in lung disease? Respir. Res. 2000, 1, 67–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhillon, S.S.; Mastropaolo, L.A.; Murchie, R.; Griffiths, C.; Thöni, C.; Elkadri, A.; Xu, W.; Mack, A.; Walters, T.; Guo, C.; et al. Higher activity of the inducible nitric oxide synthase contributes to very early onset inflammatory bowel disease. Clin. Transl. Gastroenterol. 2014, 5, e46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasbohm, E.; Fischer, S.; Küntzel, A.; Oertel, P.; Bergmann, A.; Trefz, P.; Miekisch, W.; Schubert, J.K.; Reinhold, P.; Ziller, M.; et al. Strategies for the identification of disease-related patterns of volatile organic compounds: Prediction of paratuberculosis in an animal model using random forests. J. Breath Res. 2017, 11, 047105. [Google Scholar] [CrossRef] [PubMed]
- Durán-Acevedo, C.M.; Jaimes-Mogollón, A.L.; Gualdrón-Guerrero, O.E.; Welearegay, T.G.; Martinez-Marín, J.D.; Caceres-Tarazona, J.M.; Sánchez-Acevedo, Z.C.; Beleño-Saenz, K.J.; Cindemir, U.; Österlund, L.; et al. Exhaled breath analysis for gastric cancer diagnosis in Colombian patients. Oncotarget 2018, 9, 28805–28817. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Q.; Broza, Y.Y.; Ionsecu, R.; Tisch, U.; Ding, L.; Liu, H.; Song, Q.; Pan, Y.Y.; Xiong, F.X.; Gu, K.S.; et al. A nanomaterial-based breath test for distinguishing gastric cancer from benign gastric conditions. Br. J. Cancer 2013, 108, 941–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amal, H.; Leja, M.; Broza, Y.Y.; Tisch, U.; Funka, K.; Liepniece-Karele, I.; Skapars, R.; Xu, Z.Q.; Liu, H.; Haick, H. Geographical variation in the exhaled volatile organic compounds. J. Breath Res. 2013, 7, 047102. [Google Scholar] [CrossRef] [PubMed]
- El Manouni El Hassani, S.; Berkhout, D.J.C.; Bosch, S.; Benninga, M.A.; de Boer, N.K.H.; de Meij, T.G.J. Application of fecal volatile organic compound analysis in clinical practice: Current state and future perspectives. Chemosensors 2018, 6, 29. [Google Scholar] [CrossRef]
- Bosch, S.; El Manouni El Hassani, S.; Covington, J.A.; Wicaksono, A.N.; Bomers, M.K.; Benninga, M.A.; Mulder, C.J.J.; de Boer, N.K.H.; de Meij, T.G.J. Optimized sampling conditions for fecal volatile organic compound analysis by means of field asymmetric ion mobility spectrometry. Anal. Chem. 2018, 90, 7972–7981. [Google Scholar] [CrossRef] [PubMed]
- Kou, L.; Zhang, D.; Liu, D. A novel medical e-nose signal analysis system. Sensors 2017, 17, 402. [Google Scholar] [CrossRef] [PubMed]
- Electronic ‘Nose’ Offers Rapid Epilepsy Diagnosis. Available online: https://www.medscape.com/viewarticle/889666 (accessed on 24 August 2018).
| Disease 1 | Pathogen | Host 2 | Location | Biomarkers | Chemical Class | Molecular Structure | References |
|---|---|---|---|---|---|---|---|
| Fire blight | Erwinia amylovora | Apple | Leaves | Phenylethyl alcohol | Benzene alcohol |  | [33,34] |
| Gray mold | Botrytis cinerea | Tomato | Leaves | α-Copaene | Tricyclic sesquiterpene |  | [35] |
| Powdery mildew | Oidium neolycopersici | Tomato | Leaves | 1-Fluorododecane | Fluoro-aliphatic HC |  | [36] |
| Bovine tuberculosis | Mycobacterium bovis | Cattle | Lungs | 1,3-Dimethylbutyl cycloalkane | Cycloalkane |  | [37] |
| Infective endocarditis | Staphylococcus spp. | Human | Heart | Methanethiol | Organosulfur |  | [38,39,40,41] |
| COPD, CF | Abiotic, noninfectious | Human | Lungs | Nitrotyrosine | Tyrosine deriv. |  | [42] |
| Plant Hosts 1 | Plant Part/Location | Disease Type | Pathogens/Pests | E-nose Model | E-nose Type/Sensor No. 2 | References |
|---|---|---|---|---|---|---|
| Apple | Fruit | Post-harvest rot | Unspecified | FOX 4000 | MOS 18 | [81] |
| Leaves; shoots | Fire blight, Bacterial blast | Erwinia amylovora, Pseudomonas syringae | EOS-507C/PEN3 | MOS 6, MOS 10 | [34] | |
| Leaves | Fire blight | Erwinia amylovora | EOS-835 | MOS 6 | [74] | |
| Barley | Grain | Toxigenic infestation | Aspergillus ochraceus, Aspergillus carbonarius, Penicillium verrucosum, Fusarium. graminearum, Fusarium culmorum | VCM 422 | MOSFET 10, MOS 6, Gascard CO2 1 | [82] |
| Basswood | Wood | Bacterial wetwood | Anaerobic bacteria | Aromascan A32S | CP 32 | [83] |
| Beech, Black cherry | Wood | Bacterial wetwood | Anaerobic bacteria | Aromascan A32S | CP 32 | [84] |
| Blueberry | Fruit | Fruit rot | Botrytis cinerea, Colletotrichum gloeosporioides, Alternaria species | Cyranose 320 | CBPC 32 | [85] |
| Fruit | Quality rating | unspecified | TGS 822 | MOS 2 | [86] | |
| Corn | Grain | Aflatoxins | Aspergillus flavus | PEN 2 | MOS 10 | [87,88] |
| Toxigenic infestation | Fusarium verticillioides | EOS 835 | MOS 6 | [89] | ||
| Cotton | Bolls | Wounding | Anthonomus grandis | Cyranose 320 | CBPC 32 | [90] |
| Cottonwood | Wood | Bacterial wetwood | Clostridium spp. | Aromascan A32S | CP 32 | [10] |
| Cucumber | Leaves | Spider mite | Unidentified | Bloodhound ST214 | CP 14 | [66] |
| Elm | Wood | Bacterial wetwood | Anaerobic bacteria | Aromascan A32S | CP 32 | [83] |
| Grape | Root | Root galls | Agrobacterium vitis | PEN 3 | MOS 10 | [91,92] |
| Kiwi | Fruit | Bacterial canker | Pseudomonas syringae pv. actinidiae | EOS-507C/PEN3 | MOS 6, MOS10 | [76] |
| Fruit rot | Botrytis cinerea, Sclerotinia sclerotiorum | EOS 835 | MOS 6 | [93] | ||
| Oaks | Sapwood | Oak wilt | Ceratocystis fagacearum | Aromascan A32S | CP 32 | [10,94] |
| Roots | Root rots | Armillaria mellea, Ganoderma lucidum, Heterobasidion annosum | PEN3 | MOS 10 | [95] | |
| Bole | Wood decay | Many wood decay fungi | Aromascan A32S, LibraNose 2.1, PEN 3 | CP 32, QMB 8, MOS 10 | [10,77] | |
| Oil palm | Lower bole | Basal stem rot | Ganoderma boninense | Cyranose 320 | CBPC 32 | [62] |
| Onion | Bulbs | Sour skin | Burkholderiacepacia | Owlstone | FAIMS | [96] |
| Orange | Fruit | Citrus greening | Liberibacter asiaticus | Agilent 6890 GC | MS 1 | [97] |
| Fruit rot | Penicillium spp. | LibraNose 2.1 | QMB 8 | [98] | ||
| Ornamental palm | Crown, trunk, leaves | Wounding | Rhynchophorus ferrugineous | PEN 3 | MOS 10 | [63] |
| Pear | Fruit | Post-harvest rot | Unspecified | FOX 4000 | MOS 18 | [81] |
| Pepper | Plants | Wounding | Unidentified | Bloodhound ST214 | CP 14 | [66] |
| Pine | Wood in service | Wood decay | Serpula lacrymans | Prototype | CP 10 | [99] |
| Potato | Tubers | Storage soft rot | Ralstonia solanacearum, Clavibacter michiganensis spp. sepedonicus, Pectobacterium species | Prototype | MOS 8 | [100] |
| Ralstonia solanacearum, Clavibacter michiganensis spp. sepedonicus | PEN 3 | MOS 10 | [101] | |||
| Pectobacterium carotovorum | WOLF 4.1 | EC 9, NDIR 2 | [102] | |||
| Rice | Stalks | Wilting | Nilaparvata lugens | PEN 2 | MOS 10 | [103,104] |
| Strawberry | Fruit | Fruit rot | Botrytis, Fusarium, and Penicillium spp. | PEN 3 | MOS 10 | [105] |
| Tomato | Leaves | Powdery mildew | Oidium neolycopersici | Bloodhound ST214 | CP 14 | [66] |
| Seedling | Gray mold | Botrytis cinerea | PEN 2 | MOS 10 | [106] | |
| Wheat | Grain | Decay | Penicillium chrysogenum, Fusarium verticillioides | LibraNose 2.1 | QMB 8 | [107] |
| Fusarium spp. | Prototype | QMB 8 | [108] | |||
| Rhyzopertha dominica | PEN 2 | MOS 10 | [109] |
| Animal 1 | Disease 2 | VOC Sample Type | Pathogens/Pests/Disorder | E-nose Model | E-nose Type, Sensor No. 3 | References |
|---|---|---|---|---|---|---|
| Badgers (Eurasian) | Bovine TB | Blood serum | Mycobacterium bovis | Bloodhound BH-114 | CP 14 | [126] |
| Bats (cave-dwelling) | White-nose syndrome (WNS) | Pure culture of pathogen isolated from bat skin | Pseudogymnoascus destructans | Heracles II | GC-FID 1, MOS 100s | [127,128] |
| Catfish | Flesh off-flavor | Filleted meat | Geosmin-producing aquatic Actinomycetes | Aromascan A32S | CP 32 | [129] |
| Chicken | Salmonella contamination (SC) | Meat product | Salmonella typhimurium | Spreeta | SPRB | [130] |
| Cows (cattle) | BRD | Blood serum | Mannheimia haemolytica | Bloodhound ST-214 | CP 13 | [131] |
| Bovine TB | Blood serum | Mycobacterium bovis | Bloodhound BH-114 | CP 14 | [126] | |
| Exhaled breath | Mycobacterium bovis | NA-NOSE | GNP 6 | [37] | ||
| Hyperketonaemia | Exhaled breath, milk | Metabolic disorder | Metabolomics | MS | [132] | |
| Rats | ALF | Exhaled breath | Liver cell necrosis, ischemia | eNose | MOS 8 | [133] |
| Sheep | CM | Skin | Lucilia cuprina (fly larvae) | Prototype | MOS 6 | [134] |
| Disease Name 1 | Location 2 | VOC Sample Type | Pathogen/Disorder 2 | e-Nose Model | e-Nose Type, Sensor No. 3 | References |
|---|---|---|---|---|---|---|
| ABI | Lung | Exhaled-air cultures | Pseudomonas aeruginosa, Haemophilus influenza, Streptococcus pneumoniae, Moraxella catarrhalis, Staphylococcus aureus | Cyranose 320 | CBPC 32 | [174,175] |
| ALS | Muscles | Exhaled breath | Neurodegenerative muscular disease | Cyranose 320 | CBPC 32 | [176] |
| ARDS | Lung | Exhaled breath | Lung inflammation | Cyranose 320 | CBPC 32 | [177] |
| Arthritis | Joints | Exhaled breath | Joint inflammation | Cyranose 320 | CBPC 32 | [178] |
| Asthma | Lung | Exhaled breath | Bronchial inflammation & obstruction | Cyranose 320 | CBPC 32 | [179] |
| Lung | Exhaled breath | Bronchial inflammation & obstruction | Prototype, NIOX | QMB 8, NOS 1 | [180] | |
| Bacteriuria | Urinary tract | Urine | Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Staphylococcus aureus, Staphylococcus saprophyticus, Enterococcus faecalis | Osmetech Microbial Analyzer | CP 4 | [181] |
| BAD | Bowel | Urine | Bile acid malabsorption | Fox 4000 | MOS 18 | [182] |
| BV | Vagina | Intravaginal swab culture | Neisseria gonorrhoeae, Chlamydia trachomatis, Trichomonas vaginalis | Osmetech Microbial Analyzer | CP 4 | [183] |
| Cancer | Lung | Exhaled breath | Abnormal cell growth | Prototype | QMB 8 | [184] |
| CF | Lung | Exhaled breath | Genetic disease of the mucus and sweat glands | Cyranose 320 | CBPC 32 | [185] |
| COPD | Lung | Lung bacterial cultures | Hemophilus influenzae, Streptococcus pneumoniae, Moraxella catarrhalis, gram negative-bacilli, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus viridans, Candida species, Corynebacterium spp., Staphylococcus epidermidis | Cyranose 320 | CBPC 32 | [174] |
| CKD | Kidney | Exhaled breath | Multiple causes | Prototype | MOS 6 | [186] |
| CRC | Colon | Urine | Abnormal cell growth | WOLF | EC 8, NDIR 2, PID 1 | [187] |
| Colon | Fecal | Abnormal cell growth | Cyranose 320 | CBPC 32 | [188] | |
| Colon | Breath | Abnormal cell growth | Prototype | GNP 14 | [189] | |
| DFI | Foot | Wound bacterial cultures | Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli | Cyranose 320 | CBPC 32 | [190] |
| DM | Systemic | Exhaled breath | Insufficient insulin production | Prototype | MOS 6 | [186] |
| EI | Eye | Eye swab broth cultures | Staphylococcus aureus, Haemophilus influenzae, Streptococcus pneumoniae, Escherichia coli, Pseudomonas aeruginosa, Moraxella catarrhalis | Cyranose 320 | CBPC 32 | [191] |
| ENT | Ear, nose, & throat | Sputum swab cultures | Staphylococcus aureus, Staphylococcus epidermis, Streptococcus pneumoniae, Pseudomonas aeruginosa | Cyranose 320 | CBPC 32 | [191] |
| HNC | Head, neck | Breath | Cancer | NA-NOSE | GNP 5 | [192] |
| IBD | Intestine | Urine | Unknown cause | Owlstone | FAIMS | [193] |
| Colon | Fecal | Unknown cause | Cyranose 320 | CBPC 32 | [194] | |
| Colon | Urine | Unknown cause | WOLF | EC 8, NDIR 2, PID 1 | [186] | |
| IBS | Colon | Fecal | Gastrointestinal disorder | Prototype | GC-MOS 1 | [195] |
| Colon | Aveolar breath | Gastrointestinal disorder | V&F Airsense | IMR-MS | [196] | |
| ID | Colon | Fecal | Gastroenteritis | Prototype | GC-MOS 1 | [197] |
| IEC | Heart | Oral cavity air | Bacterial inflammation of the endocardium | Prototype | MOS 6 | [198] |
| ILD | Lung | Exhaled breath | Unknown cause | NIOX | NOS 1 | [199] |
| IPA | Lungs | Exhaled breath | Aspergillus species | Cyranose 320 | CBPC 32 | [160,200] |
| IRC | Teeth | Root canal culture | Prevotella, Porphyromonas, Fusobacterium, and Bacteroides species | Shimadzu FF-1 | MOS 10 | [201] |
| LOS | Systemic | Fecal | Bacteremia | Cyranose 320 | CBPC 32 | [202] |
| MPM | Lung | Exhaled breath | Cancer of pleura tissue caused by inhalation of asbestos fibers | Cyranose 320 | CBPC 32 | [177,203] |
| NEC | Colon | Fecal | Unknown cause | Cyranose 320 | CBPC 32 | [204] |
| PA | Joints | Exhaled breath | Inflammatory joint disease | Cyranose 320 | CBPC 32 | [178] |
| PD | Systemic | Exhaled breath | Neurodegenerative disease | Prototype | GNP, CNT 40 | [205] |
| Pneumoconiosis | Lung | Exhaled breath | Occupational inhalation of dust causing inflammation | Cyranose 320 | CBPC 32 | [206] |
| Pneumonia (bacterial) | Lung | Exhaled breath | Bacterial lung infection | Cyranose 320 | CBPC 32 | [207] |
| Lung | Exhaled breath | Bacterial lung infection | Prototype | BS | [208] | |
| PS | Lung | Exhaled breath | Systemic granulomatous disease | Cyranose 320 | CBPC 32 | [209] |
| RA | Joints | Exhaled breath | Inflammatory joint disease | Cyranose 320 | CBPC 32 | [178] |
| SI | Skin, others | Skin swab culture | Staphylococcus aureus | Cyranose 320 | CBPC 32 | [210] |
| TB | Lung | Exhaled air | Mycobacterium tuberculosis | DiagNose | MOS 12 | [157] |
| Ulcer | Stomach | Exhaled breath | Helicobacter pylori | Prototype | NH4 | [211] |
| URTI | Upper respiratory tract | Exhaled breath | Bacterial pathogens | Cyranose 320 | CBPC 32 | [212] |
| UTI | Urinary tract | Urine | Staphylococcus saprophyticus, Escherichia coli, Enterococcus faecalis, Klebsiella species | ChemPro 100i | IMS 8, MOS 6 | [213] |
| VAP | Lung | Exhaled air | Bacterial pathogens | Cyranose 320 | CBPC 32 | [207,214] |
| Lung | Exhaled breath | Bacterial pathogens | DiagNose | MOS 3 | [215] | |
| WI | Skin | Skin wound cultures | Staphylococcus aureus, Staphylococcus epidermis, Streptococcus pyogenes, Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa | Prototype | MOS 6, EC 1 | [216] |
| Skin | Skin wound cultures | Staphylococcus aureus (MSSA), methicillin-resistant Staphylococcus aureus (MRSA), Streptococcus pyogenes, Escherichia coli, Pseudomonas aeruginosa, Clostridium perfringens | ChemPro 100i | IMS 8, MOS 6 | [217] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, A.D. Applications of Electronic-Nose Technologies for Noninvasive Early Detection of Plant, Animal and Human Diseases. Chemosensors 2018, 6, 45. https://0-doi-org.brum.beds.ac.uk/10.3390/chemosensors6040045
Wilson AD. Applications of Electronic-Nose Technologies for Noninvasive Early Detection of Plant, Animal and Human Diseases. Chemosensors. 2018; 6(4):45. https://0-doi-org.brum.beds.ac.uk/10.3390/chemosensors6040045
Chicago/Turabian StyleWilson, Alphus Dan. 2018. "Applications of Electronic-Nose Technologies for Noninvasive Early Detection of Plant, Animal and Human Diseases" Chemosensors 6, no. 4: 45. https://0-doi-org.brum.beds.ac.uk/10.3390/chemosensors6040045