Programming of Cell Resistance to Genotoxic and Oxidative Stress
Abstract
:1. Introduction
2. The Diversity of Mechanisms of Stress Resistance in Cancer Cells
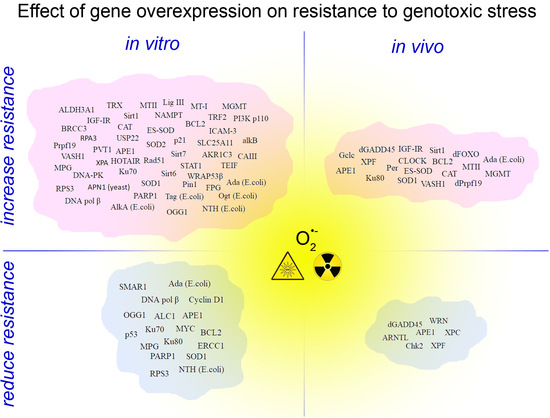
3. Genotoxic Stress Resistance in Experimental Models with Gene Overexpression
4. Prospects
Acknowledgments
Conflicts of Interest
References
- Slupphaug, G.; Kavli, B.; Krokan, H.E. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res. 2003, 531, 231–251. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Rafferty, J.A.; Hickson, I.; Chinnasamy, N.; Lashford, L.S.; Margison, G.P.; Dexter, T.M.; Fairbairn, L.J. Chemoprotection of normal tissues by transfer of drug resistance genes. Cancer Metastasis Rev. 1996, 15, 365–383. [Google Scholar] [CrossRef] [PubMed]
- Allay, J.A.; Koç, O.N.; Davis, B.M.; Gerson, S.L. Retroviral-mediated gene transduction of human alkyltransferase complementary DNA confers nitrosourea resistance to human hematopoietic progenitors. Clin. Cancer Res. 1996, 2, 1353–1359. [Google Scholar] [PubMed]
- Hickson, I.; Fairbairn, L.J.; Chinnasamy, N.; Dexter, T.M.; Margison, G.P.; Rafferty, J.A. Protection of mammalian cells against chloroethylating agent toxicity by an O6-benzylguanine-resistant mutant of human O6-alkylguanine-DNA alkyltransferase. Gene Ther. 1996, 3, 868–877. [Google Scholar] [PubMed]
- Frosina, G. Gene prophylaxis by a DNA repair function. Mol. Asp. Med. 2007, 28, 323–344. [Google Scholar] [CrossRef] [PubMed]
- Moskalev, A.A.; Shaposhnikov, M.V.; Plyusnina, E.N.; Zhavoronkov, A.; Budovsky, A.; Yanai, H.; Fraifeld, V.E. The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res. Rev. 2013, 12, 661–684. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, A.V.; Krasnov, G.S.; Dmitriev, A.A.; Alekseev, B.Y.; Kardymon, O.L.; Sadritdinova, A.F.; Fedorova, M.S.; Pokrovsky, A.V.; Melnikova, N.V.; Kaprin, A.D.; et al. Mitochondrial dysfunction and oxidative stress in aging and cancer. Oncotarget 2016, 7, 44879–44905. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, N.A.; Prakash, C.S.; McHughen, A.G. Genome editing for crop improvement: Challenges and opportunities. GM Crops Food 2015, 6, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A.; Kim, M.-H.Y.; Chappell, L.J.; Huff, J.L. How Safe Is Safe Enough? Radiation Risk for a Human Mission to Mars. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Voglauer, R.; Chang, M.W.-F.; Dampier, B.; Wieser, M.; Baumann, K.; Sterovsky, T.; Schreiber, M.; Katinger, H.; Grillari, J. SNEV overexpression extends the life span of human endothelial cells. Exp. Cell Res. 2006, 312, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Guo, J.; Yu, Y.; Yi, S.; Yu, T.; Fu, L.; Hou, L.; Chen, W. Overexpression of survivin and cyclin D1 in CHO cells confers apoptosis resistance and enhances growth in serum-free suspension culture. Biotechnol. Lett. 2011, 33, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Maeder, M.L.; Linder, S.J.; Cascio, V.M.; Fu, Y.; Ho, Q.H.; Joung, J.K. CRISPR RNA—Guided activation of endogenous human genes. Nat. Methods 2013, 10, 977–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.R.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Hilton, I.B.; D’Ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Klann, T.S.; Black, J.B.; Chellappan, M.; Safi, A.; Song, L.; Hilton, I.B.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. CRISPR–Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 2017, 35, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bloj, B.; Moses, C.; Sgro, A.; Plani-Lam, J.; Arooj, M.; Duffy, C.; Thiruvengadam, S.; Sorolla, A.; Rashwan, R.; Mancera, R.L.; et al. Waking up dormant tumor suppressor genes with zinc fingers, TALEs and the CRISPR/dCas9 system. Oncotarget 2016, 7, 60535–60554. [Google Scholar] [CrossRef] [PubMed]
- Braun, C.J.; Bruno, P.M.; Horlbeck, M.A.; Gilbert, L.A.; Weissman, J.S.; Hemann, M.T. Versatile in vivo regulation of tumor phenotypes by dCas9-mediated transcriptional perturbation. Proc. Natl. Acad. Sci. USA 2016, 113, E3892–E3900. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.-P.; Gottesman, M.M. Mechanisms of Multidrug Resistance in Cancer. In Multi-Drug Resistance in Cancer; Zhou, J., Ed.; Humana Press: Totowa, NJ, USA, 2010; Volume 596, pp. 47–76. ISBN 978-1-60761-415-9. [Google Scholar]
- Bouwman, P.; Jonkers, J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer 2012, 12, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Al-Dimassi, S.; Abou-Antoun, T.; El-Sibai, M. Cancer cell resistance mechanisms: A mini review. Clin. Transl. Oncol. 2014, 16, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, E.J.; Stanbridge, E.J.; Gupta, S.; Gupta, A.K.; Soto, D.; Bakanauskas, V.J.; Cerniglia, G.J.; Muschel, R.J.; McKenna, W.G. Direct evidence for the contribution of activated N-ras and K-ras oncogenes to increased intrinsic radiation resistance in human tumor cell lines. Cancer Res. 2000, 60, 6597–6600. [Google Scholar] [PubMed]
- Gupta, A.K.; Bakanauskas, V.J.; Cerniglia, G.J.; Cheng, Y.; Bernhard, E.J.; Muschel, R.J.; McKenna, W.G. The Ras radiation resistance pathway. Cancer Res. 2001, 61, 4278–4282. [Google Scholar] [PubMed]
- De Bacco, F.; Luraghi, P.; Medico, E.; Reato, G.; Girolami, F.; Perera, T.; Gabriele, P.; Comoglio, P.M.; Boccaccio, C. Induction of MET by Ionizing Radiation and Its Role in Radioresistance and Invasive Growth of Cancer. JNCI J. Natl. Cancer Inst. 2011, 103, 645–661. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-L, A.; Squatrito, M.; Northcott, P.; Awan, A.; Holland, E.C.; Taylor, M.D.; Nahlé, Z.; Kenney, A.M. Oncogenic YAP promotes radioresistance and genomic instability in medulloblastoma through IGF2-mediated Akt activation. Oncogene 2012, 31, 1923–1937. [Google Scholar] [CrossRef] [PubMed]
- Ghisolfi, L.; Keates, A.C.; Hu, X.; Lee, D.; Li, C.J. Ionizing radiation induces stemness in cancer cells. PLoS ONE 2012, 7, e43628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koike, K.; Abe, T.; Hisano, T.; Kubo, T.; Wada, M.; Kohno, K.; Kuwano, M. Overexpression of multidrug resistance protein gene in human cancer cell lines selected for drug resistance to epipodophyllotoxins. Jpn. J. Cancer Res. Gann 1996, 87, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, A.M.; Ambudkar, S.V. Molecular Mechanisms of Drug Resistance in Single-Step and Multi-Step Drug-Selected Cancer Cells. In Multi-Drug Resistance in Cancer; Zhou, J., Ed.; Humana Press: Totowa, NJ, USA, 2010; Volume 596, pp. 77–93. ISBN 978-1-60761-415-9. [Google Scholar]
- Breen, L.; Keenan, J.; Clynes, M. Generation of lung cancer cell line variants by drug selection or cloning. Methods Mol. Biol. 2011, 731, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Pisco, A.O.; Brock, A.; Zhou, J.; Moor, A.; Mojtahedi, M.; Jackson, D.; Huang, S. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Klein, H.L. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair 2008, 7, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.P.; Hamilton, T.C.; Schilder, R.J. Platinum Resistance: The Role of DNA Repair Pathways. Clin. Cancer Res. 2008, 14, 1291–1295. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Park, J.-M.; Yi, J.M.; Leem, S.-H.; Kang, T.-H. Enhanced nucleotide excision repair capacity in lung cancer cells by preconditioning with DNA-damaging agents. Oncotarget 2015, 6, 22575. [Google Scholar] [CrossRef] [PubMed]
- Hur, G.-C.; Cho, S.J.; Kim, C.-H.; Kim, M.K.; Bae, S.I.; Nam, S.Y.; Park, J.-W.; Kim, W.H.; Lee, B.L. Manganese superoxide dismutase expression correlates with chemosensitivity in human gastric cancer cell lines. Clin. Cancer Res. 2003, 9, 5768–5775. [Google Scholar] [PubMed]
- Zhang, D.J.; Xiang, J.; Wang, X.; Wang, J.; Xiao, J.C.; Xu, W.; Xu, H.; Xin, Y.; Zhang, L.Z.; Pei, D.S.; et al. RPA1 expression in esophageal carcinoma and its influence on radiosensitivity of esophageal carcinoma TE-1 cells. Panminerva Med. 2015, 57, 183–189. [Google Scholar] [PubMed]
- Kitahara, O.; Katagiri, T.; Tsunoda, T.; Harima, Y.; Nakamura, Y. Classification of sensitivity or resistance of cervical cancers to ionizing radiation according to expression profiles of 62 genes selected by cDNA microarray analysis. Neoplasia 2002, 4, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.M.; Kim, B.-G.; Park, C.-S.; Huh, S.J.; Kim, J.; Park, J.K.; Cho, S.M.; Kim, B.S.; Kim, J.S.; Yoo, Y.D.; et al. Increased expression of ICAM-3 is associated with radiation resistance in cervical cancer. Int. J. Cancer 2005, 117, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Sakakura, C.; Miyagawa, K.; Kuriu, Y.; Kin, S.; Nakase, Y.; Hagiwara, A.; Mitsufuji, S.; Okazaki, Y.; Hayashizaki, Y.; et al. Differential gene expression profiles of radioresistant oesophageal cancer cell lines established by continuous fractionated irradiation. Br. J. Cancer 2004, 91, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.-F.; Lin, R.-X.; Huang, J.; Zhou, Z.; Yang, J.; Guo, G.-Z.; Wang, S.-Q. Identification of differentially expressed genes contributing to radioresistance in lung cancer cells using microarray analysis. Radiat. Res. 2005, 164, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Hanna, E.; Shrieve, D.C.; Ratanatharathorn, V.; Xia, X.; Breau, R.; Suen, J.; Li, S. A novel alternative approach for prediction of radiation response of squamous cell carcinoma of head and neck. Cancer Res. 2001, 61, 2376–2380. [Google Scholar] [PubMed]
- Guo, Y.; Zhu, X.-D.; Qu, S.; Li, L.; Su, F.; Li, Y.; Huang, S.-T.; Li, D.-R. Identification of genes involved in radioresistance of nasopharyngeal carcinoma by integrating gene ontology and protein-protein interaction networks. Int. J. Oncol. 2012, 40, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Khodarev, N.N.; Beckett, M.; Labay, E.; Darga, T.; Roizman, B.; Weichselbaum, R.R. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1714–1719. [Google Scholar] [CrossRef] [PubMed]
- Souchek, J.J.; Baine, M.J.; Lin, C.; Rachagani, S.; Gupta, S.; Kaur, S.; Lester, K.; Zheng, D.; Chen, S.; Smith, L.; et al. Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Br. J. Cancer 2014, 111, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Higo, M.; Uzawa, K.; Kouzu, Y.; Bukawa, H.; Nimura, Y.; Seki, N.; Tanzawa, H. Identification of candidate radioresistant genes in human squamous cell carcinoma cells through gene expression analysis using DNA microarrays. Oncol. Rep. 2005, 14, 1293–1298. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Utsunomiya, T.; Mimori, K.; Tanaka, F.; Haraguchi, N.; Inoue, H.; Murayama, S.; Mori, M. Differential gene expression profiles of radioresistant pancreatic cancer cell lines established by fractionated irradiation. Int. J. Oncol. 2006, 28, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Sevilya, Z.; Leitner-Dagan, Y.; Pinchev, M.; Kremer, R.; Elinger, D.; Rennert, H.S.; Schechtman, E.; Freedman, L.S.; Rennert, G.; Paz-Elizur, T.; et al. Low Integrated DNA Repair Score and Lung Cancer Risk. Cancer Prev. Res. 2014, 7, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Gregel, C.M.; Kaina, B. The DNA repair protein O6-methylguanine-DNA methyltransferase protects against skin tumor formation induced by antineoplastic chloroethylnitrosourea. Cancer Res. 1997, 57, 3335–3338. [Google Scholar] [PubMed]
- Sekiguchi, M.; Nakabeppu, Y.; Sakumi, K.; Tuzuki, T. DNA-repair methyltransferase as a molecular device for preventing mutation and cancer. J. Cancer Res. Clin. Oncol. 1996, 122, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.H.; Pretlow, T.P.; O’Riordan, M.A.; Dumenco, L.L.; Allay, E.; Gerson, S.L. Transgenic expression of human MGMT protects against azoxymethane-induced aberrant crypt foci and G to A mutations in the K-ras oncogene of mouse colon. Carcinogenesis 1995, 16, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Qin, X.; Gerson, S.L. Reduced lung tumorigenesis in human methylguanine DNA—Methyltransferase transgenic mice achieved by expression of transgene within the target cell. Carcinogenesis 1999, 20, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Allay, E.; Reese, J.S.; McGuire, E.A.; Koc, O.N.; Sedransk, N.; Gerson, S.L. Potentiation of lymphomagenesis by methylnitrosourea in mice transgenic for LMO 1 is blocked by O6-alkylguanine DNA-alkyltransferase. Oncogene 1997, 15, 2127–2132. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhou, H.; Liu, L.; Gerson, S.L. Transgenic expression of human MGMT blocks the hypersensitivity of PMS2-deficient mice to low dose MNU thymic lymphomagenesis. Carcinogenesis 1999, 20, 1667–1673. [Google Scholar] [CrossRef] [PubMed]
- Canitrot, Y.; Cazaux, C.; Fréchet, M.; Bouayadi, K.; Lesca, C.; Salles, B.; Hoffmann, J.-S. Overexpression of DNA polymerase β in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl. Acad. Sci. USA 1998, 95, 12586–12590. [Google Scholar] [CrossRef] [PubMed]
- Canitrot, Y.; Frechet, M.; Servant, L.; Cazaux, C.; Hoffmann, J.S. Overexpression of DNA polymerase beta: A genomic instability enhancer process. FASEB J. 1999, 13, 1107–1111. [Google Scholar] [PubMed]
- Chan, K.; Houlbrook, S.; Zhang, Q.-M.; Harrison, M.; Hickson, I.D.; Dianov, G.L. Overexpression of DNA polymerase results in an increased rate of frameshift mutations during base excision repair. Mutagenesis 2007, 22, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Lai, Y.; Liu, S.; Wu, M.; Liu, Y.; Zhang, Z. Deregulated expression of DNA polymerase β is involved in the progression of genomic instability. Environ. Mol. Mutagen. 2012, 53, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Okuda, Y.; Nishi, R.; Ng, J.M.Y.; Vermeulen, W.; van der Horst, G.T.J.; Mori, T.; Hoeijmakers, J.H.J.; Hanaoka, F.; Sugasawa, K. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair 2004, 3, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Fortelny, N.; Overall, C.M.; Pavlidis, P.; Freue, G.V.C. Can we predict protein from mRNA levels? Nature 2017, 547, E19–E20. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.D.; Kim, T.-S.; Joo, Y.J.; Shin, H.-S.; Kim, S.-H.; Jang, C.-Y.; Lee, C.E.; Kim, J. RpS3 translation is repressed by interaction with its own mRNA. J. Cell. Biochem. 2010. [Google Scholar] [CrossRef] [PubMed]
- Glassner, B.J.; Rasmussen, L.J.; Najarian, M.T.; Posnick, L.M.; Samson, L.D. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc. Natl. Acad. Sci. USA 1998, 95, 9997–10002. [Google Scholar] [CrossRef] [PubMed]
- Coquerelle, T.; Dosch, J.; Kaina, B. Overexpression of N-methylpurine-DNA glycosylase in Chinese hamster ovary cells renders them more sensitive to the production of chromosomal aberrations by methylating agents—A case of imbalanced DNA repair. Mutat. Res. Repair 1995, 336, 9–17. [Google Scholar] [CrossRef]
- Schild, L.J.; Brookman, K.W.; Thompson, L.H.; Wilson, D.M. Effects of Ape1 overexpression on cellular resistance to DNA-damaging and anticancer agents. Somat. Cell Mol. Genet. 1999, 25, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Alamo, M.J.; Laval, F. Overexpression of the human HAP1 protein sensitizes cells to the lethal effect of bioreductive drugs. Carcinogenesis 1999, 20, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-C.; Wang, Y.-H.; Liu, Y.-C. Overexpression of PCNA Attenuates Oxidative Stress-Caused Delay of Gap-Filling during Repair of UV-Induced DNA Damage. J. Nucleic Acids 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Muruganujan, A.; Mi, H.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551. [Google Scholar] [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [PubMed]
- Pegram, M.D.; Finn, R.S.; Arzoo, K.; Beryt, M.; Pietras, R.J.; Slamon, D.J. The effect of HER-2/neu overexpression on chemotherapeutic drug sensitivity in human breast and ovarian cancer cells. Oncogene 1997, 15, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A.; Lindsey-Boltz, L.A.; Ünsal-Kaçmaz, K.; Linn, S. Molecular Mechanisms of Mammalian DNA Repair and the DNA Damage Checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Chang, L.; Sun, Y.; Hou, Y.; Fan, X.; Sun, Y. Multiplexed sgRNA Expression Allows Versatile Single Nonrepetitive DNA Labeling and Endogenous Gene Regulation. ACS Synth. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Zhao, Y.; Feng, G.; Chen, C.; Tao, Y.; Zhou, S.; Liu, S.; Chang, H.; Zeng, M.; Xia, Y. RPA3 is a potential marker of prognosis and radioresistance for nasopharyngeal carcinoma. J. Cell. Mol. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cleaver, J.E.; Charles, W.C.; McDowell, M.L.; Sadinski, W.J.; Mitchell, D.L. Overexpression of the XPA repair gene increases resistance to ultraviolet radiation in human cells by selective repair of DNA damage. Cancer Res. 1995, 55, 6152–6160. [Google Scholar] [PubMed]
- Tomicic, M.; Eschbach, E.; Kaina, B. Expression of yeast but not human apurinic/apyrimidinic endonuclease renders Chinese hamster cells more resistant to DNA damaging agents. Mutat. Res. Repair 1997, 383, 155–165. [Google Scholar] [CrossRef]
- Herring, C.J.; Deans, B.; Elder, R.H.; Rafferty, J.A.; MacKinnon, J.; Barzilay, G.; Hickson, I.D.; Hendry, J.H.; Margison, G.P. Expression levels of the DNA repair enzyme HAP1 do not correlate with the radiosensitivities of human or HAP1-transfected rat cell lines. Br. J. Cancer 1999, 80, 940. [Google Scholar] [CrossRef] [PubMed]
- Sossou, M. APE1 overexpression in XRCC1-deficient cells complements the defective repair of oxidative single strand breaks but increases genomic instability. Nucleic Acids Res. 2005, 33, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Hansen, W.K.; Deutsch, W.A.; Yacoub, A.; Xu, Y.; Williams, D.A.; Kelley, M.R. Creation of a Fully Functional Human Chimeric DNA Repair Protein COMBINING O6-Methylguanine DNA Methyltransferase (MGMT) and AP Endonuclease (APE/Redoxeffector Factor 1 (Ref 1)) DNA Repair Proteins. J. Biol. Chem. 1998, 273, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Qi, D.; Hu, Y.; Zhang, Y.; Peng, T.; Ji, W. Effect of Ku70 expression on radiosensitivity in renal carcinoma 786-O cells. Cancer Cell Int. 2014, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Kasten, U.; Borgmann, K.; Burgmann, P.; Li, G.; Dikomey, E. Overexpression of Human Ku70/Ku80 in Rat Cells Resulting in Reduced DSB Repair Capacity with Appropriate Increase in Cell Radiosensitivity but with No Effect on Cell Recovery. Radiat. Res. 1999, 151, 532. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Schultz, M.; Kruh, G.D.; Tew, K.D. Increased expression of DNA-dependent protein kinase confers resistance to adriamycin. Biochim. Biophys. Acta 1998, 1381, 131–138. [Google Scholar] [CrossRef]
- Vispé, S.; Cazaux, C.; Lesca, C.; Defais, M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998, 26, 2859–2864. [Google Scholar] [CrossRef] [PubMed]
- Rukść, A.; Birmingham, E.C.; Baker, M.D. Altered DNA repair and recombination responses in mouse cells expressing wildtype or mutant forms of RAD51. DNA Repair 2007, 6, 1876–1889. [Google Scholar] [CrossRef] [PubMed]
- Lundin, C.; Schultz, N.; Arnaudeau, C.; Mohindra, A.; Hansen, L.T.; Helleday, T. RAD51 is Involved in Repair of Damage Associated with DNA Replication in Mammalian Cells. J. Mol. Biol. 2003, 328, 521–535. [Google Scholar] [CrossRef]
- Ahel, D.; Horejsi, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P.; et al. Poly(ADP-ribose)-Dependent Regulation of DNA Repair by the Chromatin Remodeling Enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar] [CrossRef] [PubMed]
- Ho, E.L.Y. Repair of single-strand DNA interruptions by redundant pathways and its implication in cellular sensitivity to DNA-damaging agents. Nucleic Acids Res. 2003, 31, 7032–7040. [Google Scholar] [CrossRef] [PubMed]
- Klungland, A.; Fairbairn, L.; Watson, A.J.; Margison, G.P.; Seeberg, E. Expression of the E. coli 3-methyladenine DNA glycosylase I gene in mammalian cells reduces the toxic and mutagenic effects of methylating agents. EMBO J. 1992, 11, 4439. [Google Scholar] [PubMed]
- Klungland, A.; Bjørås, M.; Hoff, E.; Seeberg, E. Increased removal of 3-alkyladenine reduces the frequencies of hprt mutations induced by methyl- and ethylmethanesulfonate in Chinese hamster fibroblast cells. Nucleic Acids Res. 1994, 22, 1670–1674. [Google Scholar] [CrossRef] [PubMed]
- Imperatori, L.; Damia, G.; Taverna, P.; Garattini, E.; Citti, L.; Boldrini, L.; D’Incalci, M. 3T3 NIH murine fibroblasts and B78 murine melanoma cells expressing the Escherichia coli N3-methyladenine-DNA glycosylase I do not become resistant to alkylating agents. Carcinogenesis 1994, 15, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Habraken, Y.; Laval, F. Increased resistance of the Chinese hamster mutant irsl cells to monofunctional alkylating agents by transfection of the E. coli or mammalian N3-methyladenine-DNA-glycosylase genes. Mutat. Res. Repair 1993, 293, 187–195. [Google Scholar] [CrossRef]
- Calléja, F.; Jansen, J.G.; Vrieling, H.; Laval, F.; van Zeeland, A.A. Modulation of the toxic and mutagenic effects induced by methyl methanesulfonate in Chinese hamster ovary cells by overexpression of the rat N-alkylpurine-DNA glycosylase. Mutat. Res. Mol. Mech. Mutagen. 1999, 425, 185–194. [Google Scholar] [CrossRef]
- Bramson, J.; O’Connor, T.; Panasci, L. Effect of alkyl-N-purine DNA glycosylase overexpression on cellular resistance to bifunctional alkylating agents. Biochem. Pharmacol. 1995, 50, 39–44. [Google Scholar] [CrossRef]
- Ibeanu, G.; Hartenstein, B.; Dunn, W.C.; Chang, L.-Y.; Hofmann, E.; Coquerelle, T.; Mitra, S.; Kaina, B. Overexpression of human DNA repair protein N-methylpurine-DNA glycosylase results in the increased removal of N-methylpurines in DNA without a concomitant increase in resistance to alkylating agents in Chinese hamster ovary cells. Carcinogenesis 1992, 13, 1989–1995. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, R.N.; Almeida, K.H.; Fornsaglio, J.L.; Schamus, S.; Sobol, R.W. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res. 2005, 65, 6394–6400. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, R.N.; Wang, X.-H.; Jelezcova, E.; Goellner, E.M.; Tang, J.-B.; Sobol, R.W. Human Methyl Purine DNA Glycosylase and DNA Polymerase Expression Collectively Predict Sensitivity to Temozolomide. Mol. Pharmacol. 2008, 74, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Laval, F. Expression of the E. coli fpg gene in mammalian cells reduces the mutagenicity of gamma-rays. Nucleic Acids Res. 1994, 22, 4943–4946. [Google Scholar] [CrossRef] [PubMed]
- Cussac, C.; Laval, F. Reduction of the toxicity and mutagenicity of aziridine in mammalian cells harboring the Escherichia coli fpg gene. Nucleic Acids Res. 1996, 24, 1742–1746. [Google Scholar] [CrossRef] [PubMed]
- Radyuk, S.N.; Michalak, K.; Rebrin, I.; Sohal, R.S.; Orr, W.C. Effects of ectopic expression of Drosophila DNA glycosylases dOgg1 and RpS3 in mitochondria. Free Radic. Biol. Med. 2006, 41, 757–764. [Google Scholar] [CrossRef] [PubMed]
- Hollenbach, S.; Dhénaut, A.; Eckert, I.; Radicella, J.P.; Epe, B. Overexpression of Ogg1 in mammalian cells: Effects on induced and spontaneous oxidative DNA damage and mutagenesis. Carcinogenesis 1999, 20, 1863–1868. [Google Scholar] [CrossRef] [PubMed]
- Bramson, J.; Panasci, L.C. Effect of ERCC-1 overexpression on sensitivity of Chinese hamster ovary cells to DNA damaging agents. Cancer Res. 1993, 53, 3237–3240. [Google Scholar] [PubMed]
- Harrison, L.; Skorvaga, M.; Cunningham, R.P.; Hendry, J.H.; Margison, G.P. Transfection of the Escherichia coli nth Gene into Radiosensitive Chinese Hamster Cells: Effects on Sensitivity to Radiation, Hydrogen Peroxide, and Bleomycin Sulfate. Radiat. Res. 1992, 132, 30. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.C.; Margison, G.P. Expression in mammalian cells of the Escherichia coli O6 alkylguanine-DNA-alkyltransferase gene ogt reduces the toxicity of alkylnitrosoureas. Br. J. Cancer 1993, 67, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Abril, N.; Margison, G.P. Mammalian Cells Expressing Escherichia coli O6-Alkylguanine-DNA Alkyltransferases Are Hypersensitive to Dibromoalkanes. Chem. Res. Toxicol. 1999, 12, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Von Hofe, E.; Fairbairn, L.; Margison, G.P. Relationship between O6-alkylguanine-DNA alkyltransferase activity and N-methyl-N’-nitro-N-nitrosoguanidine-induced mutation, transformation, and cytotoxicity in C3H/10T1/2 cells expressing exogenous alkyltransferase genes. Proc. Natl. Acad. Sci. USA 1992, 89, 11199–11203. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, K.; Tsujimura, T.; Yawata, H.; Fujio, C.; Nakabeppu, Y.; Sekiguchi, M.; Ikenaga, M. Transfer of the E. coli O6-methyltransferase gene into repair-deficient human cells and restoration of cellular resistance to N-methyl-N′-nitro-N-nitrosoguanidine. Mutat. Res. Repair Rep. 1986, 166, 135–141. [Google Scholar] [CrossRef]
- Ishizaki, K.; Tsujimura, T.; Fujio, C.; Zhang, Y.P.; Yawata, H.; Nakabeppu, Y.; Sekiguchi, M.; Ikenaga, M. Expression of the truncated E. coli O6-methylguanine methyltransferase gene in repair-deficient human cells and restoration of cellular resistance to alkylating agents. Mutat. Res. 1987, 184, 121–128. [Google Scholar] [PubMed]
- Dumenco, L.L.; Warman, B.; Hatzoglou, M.; Lim, I.K.; Abboud, S.L.; Gerson, S.L. Increase in nitrosourea resistance in mammalian cells by retrovirally mediated gene transfer of bacterial O6-alkylguanine-DNA alkyltransferase. Cancer Res. 1989, 49, 6044–6051. [Google Scholar] [PubMed]
- Lim, I.K.; Dumenco, L.L.; Hatzoglou, M.; Hanson, R.W.; Gerson, S.L. Increased drug resistance following retroviral gene transfer of a chimeric P-enolpyruvate carboxykinase (GTP)-bacterial O6-alkylguanine-DNA alkyltransferase gene into NRK cells. Carcinogenesis 1990, 11, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Jelinek, J.; Kleibl, K.; Dexter, T.M.; Margison, G.P. Transfection of murine multi-potent haemopoietic stem cells with an E. coli DNA alkyltransferase gene confers resistance to the toxic effects of alkylating agents. Carcinogenesis 1988, 9, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Kaina, B.; Fritz, G.; Coquerelle, T. Identification of human genes involved in repair and tolerance of DNA damage. Radiat. Environ. Biophys. 1991, 30, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, H.; Hall, J.; Karran, P. Complementation of sensitivity to alkylating agents in Escherichia coli and Chinese hamster ovary cells by expression of a cloned bacterial DNA repair gene. EMBO J. 1986, 5, 3195. [Google Scholar] [PubMed]
- Brennand, J.; Margison, G.P. Expression in mammalian cells of a truncated Escherichia coli gene coding for O6-alkylguanine alkyltransferase reduces the toxic effects of alkylating agents. Carcinogenesis 1986, 7, 2081–2084. [Google Scholar] [CrossRef] [PubMed]
- Brennand, J.; Margison, G.P. Reduction of the toxicity and mutagenicity of alkylating agents in mammalian cells harboring the Escherichia coli alkyltransferase gene. Proc. Natl. Acad. Sci. USA 1986, 83, 6292–6296. [Google Scholar] [CrossRef] [PubMed]
- White, G.R.; Ockey, C.H.; Brennand, J.; Margison, G.P. Chinese hamster cells harbouring the Escherichia coli O6-alkylguanine alkyltransferase gene are less susceptible to sister chromatid exchange induction and chromosome damage by methylating agents. Carcinogenesis 1986, 7, 2077–2080. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.; Brennand, J.; Margison, G.P. Protection of Chinese hamster cells against the cytotoxic and mutagenic effects of alkylating agents by transfection of the Escherichia coli alkyltransferase gene and a truncated derivative. Mutagenesis 1987, 2, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Minnick, D.T.; Gerson, S.L.; Dumenco, L.L.; Veigl, M.L.; Sedwick, W.D. Specificity of bischloroethylnitrosourea-induced mutation in a Chinese hamster ovary cell line transformed to express human O6-alkylguanine-DNA alkyltransferase. Cancer Res. 1993, 53, 997–1003. [Google Scholar] [PubMed]
- Allay, J.A.; Dumenco, L.L.; Koc, O.N.; Liu, L.; Gerson, S.L. Retroviral transduction and expression of the human alkyltransferase cDNA provides nitrosourea resistance to hematopoietic cells. Blood 1995, 85, 3342–3351. [Google Scholar] [PubMed]
- Moritz, T.; Mackay, W.; Glassner, B.J.; Williams, D.A.; Samson, L. Retrovirus-mediated expression of a DNA repair protein in bone marrow protects hematopoietic cells from nitrosourea-induced toxicity in vitro and in vivo. Cancer Res. 1995, 55, 2608–2614. [Google Scholar] [PubMed]
- Reese, J.S.; Koç, O.N.; Lee, K.M.; Liu, L.; Allay, J.A.; Phillips, W.P.; Gerson, S.L. Retroviral transduction of a mutant methylguanine DNA methyltransferase gene into human CD34 cells confers resistance to O6-benzylguanine plus 1,3-bis(2-chloroethyl)-1-nitrosourea. Proc. Natl. Acad. Sci. USA 1996, 93, 14088–14093. [Google Scholar] [CrossRef] [PubMed]
- Jelinek, J.; Fairbairn, L.J.; Dexter, T.M.; Rafferty, J.A.; Stocking, C.; Ostertag, W.; Margison, G.P. Long-term protection of hematopoiesis against the cytotoxic effects of multiple doses of nitrosourea by retrovirus-mediated expression of human O6-alkylguanine-DNA-alkyltransferase. Blood 1996, 87, 1957–1961. [Google Scholar] [PubMed]
- Reese, J.S.; Davis, B.M.; Liu, L.; Gerson, S.L. Simultaneous protection of G156A methylguanine DNA methyltransferase gene-transduced hematopoietic progenitors and sensitization of tumor cells using O6-benzylguanine and temozolomide. Clin. Cancer Res. 1999, 5, 163–169. [Google Scholar] [PubMed]
- Maze, R.; Carney, J.P.; Kelley, M.R.; Glassner, B.J.; Williams, D.A.; Samson, L. Increasing DNA repair methyltransferase levels via bone marrow stem cell transduction rescues mice from the toxic effects of 1,3-bis(2-chloroethyl)-1-nitrosourea, a chemotherapeutic alkylating agent. Proc. Natl. Acad. Sci. USA 1996, 93, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Bignami, M.; Terlizzese, M.; Zijno, A.; Calcagnile, A.; Frosina, G.; Abbondandolo, A.; Dogliotti, E. Cytotoxicity, mutations and SCEs induced by methylating agents are reduced in CHO cells expressing an active mammalian O6-methylguanine-DNA methyltransferase gene. Carcinogenesis 1987, 8, 1417–1421. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.J.; Carroll, P.; Samson, L. The Escherichia coli AlkB protein protects human cells against alkylation-induced toxicity. J. Bacteriol. 1994, 176, 6255–6261. [Google Scholar] [CrossRef] [PubMed]
- Veldwijk, M.R.; Trah, J.; Wang, M.; Maier, P.; Fruehauf, S.; Zeller, W.J.; Herskind, C.; Wenz, F. Overexpression of Manganese Superoxide Dismutase Does Not Increase Clonogenic Cell Survival Despite Effect on Apoptosis in Irradiated Lymphoblastoid Cells. Radiat. Res. 2011, 176, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Veldwijk, M.R.; Herskind, C.; Sellner, L.; Radujkovic, A.; Laufs, S.; Fruehauf, S.; Zeller, W.J.; Wenz, F. Normal-Tissue Radioprotection by Overexpression of the Copper-Zinc and Manganese Superoxide Dismutase Genes. Strahlenther. Onkol. 2009, 185, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Carlson, E.C.; Pellet, L.; Moritz, J.T.; Epstein, P.N. Overexpression of metallothionein in pancreatic β-cells reduces streptozotocin-induced DNA damage and diabetes. Diabetes 2001, 50, 2040–2046. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Anderson, C.M.; Chen, Y.; Stein, B.A.; Fahlman, C.S.; Copin, J.C.; Chan, P.H.; Swanson, R.A. Differing effects of copper, zinc superoxide dismutase overexpression on neurotoxicity elicited by nitric oxide, reactive oxygen species, and excitotoxins. J. Cereb. Blood Flow Metab. 2000, 20, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Rezvani, H.R.; Mazurier, F.; Cario-André, M.; Pain, C.; Ged, C.; Taïeb, A.; de Verneuil, H. Protective Effects of Catalase Overexpression on UVB-induced Apoptosis in Normal Human Keratinocytes. J. Biol. Chem. 2006, 281, 17999–18007. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Sarsour, E.H.; Kalen, A.L.; Li, L.; Kumar, M.G.; Goswami, P.C. Late ROS accumulation and radiosensitivity in SOD1-overexpressing human glioma cells. Free Radic. Biol. Med. 2008, 45, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-M.; Cheng, Y.-W.; Wu, T.-C.; Chen, C.-Y.; Lee, H. MnSOD overexpression confers cisplatin resistance in lung adenocarcinoma via the NF-κB/Snail/Bcl-2 pathway. Free Radic. Biol. Med. 2015, 79, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.J.; Goswami, P.C. Mitochondria-targeted antioxidant enzyme activity regulates radioresistance in human pancreatic cancer cells. Cancer Biol. Ther. 2008, 7, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Yan-Sanders, Y.; Lyn-Cook, B.D.; Wang, T.; Tamae, D.; Ogi, J.; Khaletskiy, A.; Li, Z.; Weydert, C.; Longmate, J.A.; et al. Manganese Superoxide Dismutase-Mediated Gene Expression in Radiation-Induced Adaptive Responses. Mol. Cell. Biol. 2003, 23, 2362–2378. [Google Scholar] [CrossRef] [PubMed]
- Motoori, S.; Majima, H.J.; Ebara, M.; Kato, H.; Hirai, F.; Kakinuma, S.; Yamaguchi, C.; Ozawa, T.; Nagano, T.; Tsujii, H.; et al. Overexpression of mitochondrial manganese superoxide dismutase protects against radiation-induced cell death in the human hepatocellular carcinoma cell line HLE. Cancer Res. 2001, 61, 5382–5388. [Google Scholar] [PubMed]
- Voulgaridou, G.-P.; Kiziridou, M.; Mantso, T.; Chlichlia, K.; Galanis, A.; Koukourakis, M.I.; Franco, R.; Panayiotidis, M.I.; Pappa, A. Aldehyde dehydrogenase 3A1 promotes multi-modality resistance and alters gene expression profile in human breast adenocarcinoma MCF-7 cells. Int. J. Biochem. Cell Biol. 2016, 77, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yang, H.; Ramesh, A.; Goodwin, J.S.; Okoro, E.U.; Guo, Z. Overexpression of Catalase Enhances Benzo(a)pyrene Detoxification in Endothelial Microsomes. PLoS ONE 2016, 11, e0162561. [Google Scholar] [CrossRef] [PubMed]
- Umeda-Kameyama, Y.; Tsuda, M.; Ohkura, C.; Matsuo, T.; Namba, Y.; Ohuchi, Y.; Aigaki, T. Thioredoxin Suppresses Parkin-associated Endothelin Receptor-like Receptor-induced Neurotoxicity and Extends Longevity in Drosoph. J. Biol. Chem. 2007, 282, 11180–11187. [Google Scholar] [CrossRef] [PubMed]
- Kaina, B.; Lohrer, H.; Karin, M.; Herrlich, P. Overexpressed human metallothionein IIA gene protects Chinese hamster ovary cells from killing by alkylating agents. Proc. Natl. Acad. Sci. USA 1990, 87, 2710–2714. [Google Scholar] [CrossRef] [PubMed]
- Kelley, S.L.; Basu, A.; Teicher, B.A.; Hacker, M.P.; Hamer, D.H.; Lazo, J.S. Overexpression of metallothionein confers resistance to anticancer drugs. Science 1988, 241, 1813–1815. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.A.; Lazo, J.S.; Yalowich, J.C.; Reynolds, I.; Kagan, V.E.; Tyurin, V.; Kim, Y.M.; Watkins, S.C.; Pitt, B.R. Cytoplasmic metallothionein overexpression protects NIH 3T3 cells from tert-butyl hydroperoxide toxicity. J. Biol. Chem. 1994, 269, 15238–15243. [Google Scholar] [PubMed]
- Sarkar, B. (Ed.) Genetic Response to Metals; CRC Press: Boca Raton, FL, USA, 1995; ISBN 978-0-8247-9615-0. [Google Scholar]
- Goncharova, E.I.; Rossman, T.G. A role for metallothionein and zinc in spontaneous mutagenesis. Cancer Res. 1994, 54, 5318–5323. [Google Scholar] [PubMed]
- Coco Martin, J.M.; Balkenende, A.; Verschoor, T.; Lallemand, F.; Michalides, R. Cyclin D1 overexpression enhances radiation-induced apoptosis and radiosensitivity in a breast tumor cell line. Cancer Res. 1999, 59, 1134–1140. [Google Scholar] [PubMed]
- Kokunai, T.; Tamaki, N. Relationship between expression of p21WAF1/CIP1 and radio resistance in human gliomas. Cancer Sci. 1999, 90, 638–646. [Google Scholar]
- Sentman, C.L.; Shutter, J.R.; Hockenbery, D.; Kanagawa, O.; Korsmeyer, S.J. Bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell 1991, 67, 879–888. [Google Scholar] [CrossRef]
- Pourzand, C.; Rossier, G.; Reelfs, O.; Borner, C.; Tyrrell, R.M. Overxpression of Bcl-2 inhibits UVA-mediated immediate apoptosiinrat 6 fibroblasts: Evidence for the involvement of Bcl-2 as an antioxidant. Cancer Res. 1997, 57, 1405–1411. [Google Scholar] [PubMed]
- Kong, C.-Z.; Zhang, Z. Bcl-2 Overexpression Inhibits Generation of Intracellular Reactive Oxygen Species and Blocks Adriamycin-induced Apoptosis in Bladder Cancer Cells. Asian Pac. J. Cancer Prev. 2013, 14, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Itamochi, H.; Yamasaki, F.; Sudo, T.; Takahashi, T.; Bartholomeusz, C.; Das, S.; Terakawa, N.; Ueno, N.T. Reduction of radiation-induced apoptosis by specific expression of Bcl-2 in normal cells. Cancer Gene Ther. 2006, 13, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, F.; May, W.S.; Zhang, Y.; Flagg, T.; Deng, X. Bcl2 Negatively Regulates DNA Double-Strand-Break Repair through a Nonhomologous End-Joining Pathway. Mol. Cell 2008, 29, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Ning, Z.; Lu, C.; Gao, W.; Liang, J.; Yan, Q.; Tan, G.; Liu, J. USP22 Induces Cisplatin Resistance in Lung Adenocarcinoma by Regulating γH2AX-Mediated DNA Damage Repair and Ku70/Bax-Mediated Apoptosis. Front. Pharmacol. 2017, 8, 274. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Yu, D.; Miura, M. Overexpression of IGF-I receptor in HeLa cells enhances in vivo radioresponse. Biochem. Biophys. Res. Commun. 2007, 363, 937–941. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.C.; Haffty, B.G.; Narayanan, L.; Yuan, J.; Havre, P.A.; Gumbs, A.A.; Kaplan, L.; Burgaud, J.L.; Carter, D.; Baserga, R.; et al. Insulin-like growth factor-I receptor overexpression mediates cellular radioresistance and local breast cancer recurrence after lumpectomy and radiation. Cancer Res. 1997, 57, 3079–3083. [Google Scholar] [PubMed]
- Nakamura, S.; Watanabe, H.; Miura, M.; Sasaki, T. Effect of the Insulin-like Growth Factor I Receptor on Ionizing Radiation-Induced Cell Death in Mouse Embryo Fibroblasts. Exp. Cell Res. 1997, 235, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, M.; Watanabe, H.; Nakamura, S.; Yu, D.; Aung, W.; Sasaki, T.; Shibuya, H.; Miura, M. Antiapoptotic activity is dispensable for insulin-like growth factor I receptor-mediated clonogenic radioresistance after gamma-irradiation. Clin. Cancer Res. 2001, 7, 3206–3214. [Google Scholar] [PubMed]
- Yu, D.; Watanabe, H.; Shibuya, H.; Miura, M. Redundancy of Radioresistant Signaling Pathways Originating from Insulin-like Growth Factor I Receptor. J. Biol. Chem. 2003, 278, 6702–6709. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-C.; Jeng, Y.-M.; Yuan, R.-H.; Hsu, H.-C.; Chen, Y.-L. SIRT1 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma and its expression predicts poor prognosis. Ann. Surg. Oncol. 2012, 19, 2011–2019. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.W.; Choi, Y.J.; Park, M.H.; Jang, E.J.; Kim, D.H.; Park, B.H.; Yu, B.P.; Chung, H.Y. Molecular Insights into SIRT1 Protection Against UVB-Induced Skin Fibroblast Senescence by Suppression of Oxidative Stress and p53 Acetylation. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Asaka, R.; Miyamoto, T.; Yamada, Y.; Ando, H.; Mvunta, D.H.; Kobara, H.; Shiozawa, T. Sirtuin 1 promotes the growth and cisplatin resistance of endometrial carcinoma cells: A novel therapeutic target. Lab. Investig. 2015, 95, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xia, L.; Zhang, Y.; Wang, H.; Xu, W.; Hu, H.; Wang, J.; Xin, J.; Gang, Y.; Sha, S.; et al. Activating Transcription Factor 4 Confers a Multidrug Resistance Phenotype to Gastric Cancer Cells through Transactivation of SIRT1 Expression. PLoS ONE 2012, 7, e31431. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Hine, C.; Tian, X.; Van Meter, M.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 Promotes DNA Repair Under Stress by Activating PARP1. Science 2011, 332, 1443–1446. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Hasan, M.K.; Alvarado, E.; Yuan, H.; Wu, H.; Chen, W.Y. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene 2011, 30, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, H.; Watanabe, T.; Hayashi, H.; Suzuki, Y.; Nakamura, T.; Ito, S.; Ono, M.; Hoshikawa, Y.; Okada, Y.; Kondo, T.; et al. Angiogenesis Inhibitor Vasohibin-1 Enhances Stress Resistance of Endothelial Cells via Induction of SOD2 and SIRT1. PLoS ONE 2012, 7, e46459. [Google Scholar] [CrossRef] [PubMed]
- Kiran, S.; Oddi, V.; Ramakrishna, G. Sirtuin 7 promotes cellular survival following genomic stress by attenuation of DNA damage, SAPK activation and p53 response. Exp. Cell Res. 2015, 331, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Xu, B.; Qu, C.; Tao, Y.; Chen, C.; Hua, W.; Feng, G.; Chang, H.; Liu, Z.; Li, G.; et al. BRCC3 acts as a prognostic marker in nasopharyngeal carcinoma patients treated with radiotherapy and mediates radiation resistance in vitro. Radiat. Oncol. 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Oshima, M.; Yuan, J.; Saraya, A.; Miyagi, S.; Konuma, T.; Yamazaki, S.; Osawa, M.; Nakauchi, H.; Koseki, H.; et al. Bmi1 Confers Resistance to Oxidative Stress on Hematopoietic Stem Cells. PLoS ONE 2012, 7, e36209. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, H.M.; Brock, S.; Gray, J.J.; Linseman, D.A. Stable over-expression of the 2-oxoglutarate carrier enhances neuronal cell resistance to oxidative stress via Bcl-2-dependent mitochondrial GSH transport. J. Neurochem. 2014, 130, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.-Q.; Gu, X.; Gao, X.-S.; Li, Y.; Yu, H.; Xiong, W.; Yu, H.; Wang, W.; Li, Y.; Teng, Y.; et al. Overexpression of AKR1C3 significantly enhances human prostate cancer cells resistance to radiation. Oncotarget 2016, 7, 48050–48058. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liu, Z.; Shi, F.; Wang, J. Pin1 modulates chemo-resistance by up-regulating FoxM1 and the involvements of Wnt/β-catenin signaling pathway in cervical cancer. Mol. Cell. Biochem. 2016, 413, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bu, P.; Liu, L.; Zhang, X.; Li, J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem. Biophys. Res. Commun. 2015, 462, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Liu, Z.; Zhou, Y.; Mi, R.; Wang, D. Overexpression of long non-coding RNA PVT1 in ovarian cancer cells promotes cisplatin resistance by regulating apoptotic pathways. Int. J. Clin. Exp. Med. 2015, 8, 20565–20572. [Google Scholar] [PubMed]
- Rassoolzadeh, H.; Böhm, S.; Hedström, E.; Gad, H.; Helleday, T.; Henriksson, S.; Farnebo, M. Overexpression of the scaffold WD40 protein WRAP53β enhances the repair of and cell survival from DNA double-strand breaks. Cell Death Dis. 2016, 7, e2267. [Google Scholar] [CrossRef] [PubMed]
- Richter, T.; Saretzki, G.; Nelson, G.; Melcher, M.; Olijslagers, S.; von Zglinicki, T. TRF2 overexpression diminishes repair of telomeric single-strand breaks and accelerates telomere shortening in human fibroblasts. Mech. Ageing Dev. 2007, 128, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Seluanov, A.; Jiang, Y.; Gorbunova, V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. USA 2007, 104, 13068–13073. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, A.; Deb-Basu, D.; Cherry, A.; Turner, S.; Ford, J.; Felsher, D.W. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc. Natl. Acad. Sci. USA 2003, 100, 9974–9979. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zheng, J.; Ling, Y.; Hou, L.; Zhang, B. Transcriptional upregulation of DNA polymerase β by TEIF. Biochem. Biophys. Res. Commun. 2005, 333, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Bernges, F.; Bürkle, A.; Küpper, J.-H.; Zeller, W.J. Functional overexpression of human poly (ADP-ribose) polymerase in transfected rat tumor cells. Carcinogenesis 1997, 18, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Van Gool, L.; Meyer, R.; Tobiasch, E.; Cziepluch, C.; Jauniaux, J.C.; Mincheva, A.; Lichter, P.; Poirier, G.G.; Bürkle, A.; Küpper, J.H. Overexpression of human poly(ADP-ribose) polymerase in transfected hamster cells leads to increased poly(ADP-ribosyl)ation and cellular sensitization to gamma irradiation. Eur. J. Biochem. 1997, 244, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Fritz, G.; Auer, B.; Kaina, B. Effect of transfection of human poly(ADP-ribose)polymerase in Chinese hamster cells on mutagen resistance. Mutat. Res. 1994, 308, 127–133. [Google Scholar] [CrossRef]
- Özeş, A.R.; Miller, D.F.; Özeş, O.N.; Fang, F.; Liu, Y.; Matei, D.; Huang, T.; Nephew, K.P. NF-κB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016, 35, 5350–5361. [Google Scholar] [CrossRef] [PubMed]
- Kelley, M.R.; Tritt, R.; Xu, Y.; New, S.; Freie, B.; Clapp, D.W.; Deutsch, W.A. The Drosophila S3 multifunctional DNA repair/ribosomal protein protects Fanconi anemia cells against oxidative DNA damaging agents. Mutat. Res. Repair 2001, 485, 107–119. [Google Scholar] [CrossRef]
- Choi, S.H.; Kim, S.Y.; An, J.J.; Lee, S.H.; Kim, D.W.; Ryu, H.J.; Lee, N.I.; Yeo, S.I.; Jang, S.H.; Won, M.H.; et al. Human PEP-1-ribosomal protein S3 protects against UV-induced skin cell death. FEBS Lett. 2006, 580, 6755–6762. [Google Scholar] [CrossRef] [PubMed]
- Räisänen, S.R.; Lehenkari, P.; Tasanen, M.; Rahkila, P.; Härkönen, P.L.; Väänänen, H.K. Carbonic anhydrase III protects cells from hydrogen peroxide-induced apoptosis. FASEB J. 1999, 13, 513–522. [Google Scholar] [CrossRef]
- Ye, S.; Shen, J.; Choy, E.; Yang, C.; Mankin, H.; Hornicek, F.; Duan, Z. p53 overexpression increases chemosensitivity in multidrug-resistant osteosarcoma cell lines. Cancer Chemother. Pharmacol. 2016, 77, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Yoon, S.; Park, M.S.; Kim, J.H.; Lee, J.-H.; Song, C.-W. Influence of p53 expression on sensitivity of cancer cells to bleomycin. J. Biochem. Mol. Toxicol. 2010, 24, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Guntur, V.P.; Waldrep, J.C.; Guo, J.J.; Selting, K.; Dhand, R. Increasing p53 protein sensitizes non-small cell lung cancer to paclitaxel and cisplatin in vitro. Anticancer Res. 2010, 30, 3557–3564. [Google Scholar] [PubMed]
- Zellars, R.C.; Naida, J.D.; Davis, M.A.; Lawrence, T.S. Effect of p53 overexpression on radiation sensitivity of human colon cancer cells. Radiat. Oncol. Investig. 1997, 5, 43–49. [Google Scholar] [CrossRef]
- Liu, H.; Ma, F.; Shen, Y.; Hu, Y.; Pan, S. Overexpression of SMAR1 Enhances Radiosensitivity in Human Breast Cancer Cell Line MCF7 via Activation of p53 Signaling Pathway. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2015, 22, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Shilova, L.A.; Pliusnina, E.N.; Zemskaia, N.V.; Moskalev, A.A. Role of DNA repair genes in radiation-induced changes of lifespan of Drosophila melanogaster. Radiat. Biol. Radioecol. 2014, 54, 482–492. [Google Scholar]
- Shaposhnikov, M.; Proshkina, E.; Shilova, L.; Zhavoronkov, A.; Moskalev, A. Lifespan and Stress Resistance in Drosophila with Overexpressed DNA Repair Genes. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Garschall, K.; Dellago, H.; Gáliková, M.; Schosserer, M.; Flatt, T.; Grillari, J. Ubiquitous overexpression of the DNA repair factor dPrp19 reduces DNA damage and extends Drosophila life span. NPJ Aging Mech. Dis. 2017, 3. [Google Scholar] [CrossRef]
- Nakatsuru, Y.; Matsukuma, S.; Nemoto, N.; Sugano, H.; Sekiguchi, M.; Ishikawa, T. O6-methylguanine-DNA methyltransferase protects against nitrosamine-induced hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 1993, 90, 6468–6472. [Google Scholar] [CrossRef] [PubMed]
- Dumenco, L.L.; Arce, C.; Norton, K.; Yun, J.; Wagner, T.; Gerson, S.L. Enhanced repair of O6-methylguanine DNA adducts in the liver of transgenic mice expressing the ada gene. Cancer Res. 1991, 51, 3391–3398. [Google Scholar] [PubMed]
- Davis, B.M.; Reese, J.S.; Koç, O.N.; Lee, K.; Schupp, J.E.; Gerson, S.L. Selection for G156A O6-methylguanine DNA methyltransferase gene-transduced hematopoietic progenitors and protection from lethality in mice treated with O6-benzylguanine and 1, 3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res. 1997, 57, 5093–5099. [Google Scholar] [PubMed]
- Dumenco, L.L.; Allay, E.; Norton, K.; Gerson, S.L. The prevention of thymic lymphomas in transgenic mice by human O6-alkylguanine-DNA alkyltransferase. Science 1993, 259, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Allay, E.; Dumenco, L.L.; Gerson, S.L. Rapid repair of O6-methylguanine-DNA adducts protects transgenic mice from N-methylnitrosourea-induced thymic lymphomas. Cancer Res. 1994, 54, 4648–4652. [Google Scholar] [PubMed]
- Allay, E.; Veigl, M.; Gerson, S.L. Mice over-expressing human O6 alkylguanine-DNA alkyltransferase selectively reduce O6 methylguanine mediated carcinogenic mutations to threshold levels after N-methyl-N-nitrosourea. Oncogene 1999, 18, 3783–3787. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Dosch, J.; Gregel, C.M.; Martin, B.A.; Kaina, B. Targeted expression of human O6-methylguanine-DNA methyltransferase (MGMT) in transgenic mice protects against tumor initiation in two-stage skin carcinogenesis. Cancer Res. 1996, 56, 3244–3249. [Google Scholar] [PubMed]
- Moskalev, A.; Shaposhnikov, M.; Proshkina, E.; Belyi, A.; Fedintsev, A.; Zhikrivetskaya, S.; Guvatova, Z.; Sadritdinova, A.; Snezhkina, A.; Krasnov, G.; et al. The influence of pro-longevity gene Gclc overexpression on the age-dependent changes in Drosophila transcriptome and biological functions. BMC Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Parkes, T.L.; Elia, A.J.; Dickinson, D.; Hilliker, A.J.; Phillips, J.P.; Boulianne, G.L. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat. Genet. 1998, 19, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Orr, W.C.; Sohal, R.S. Effects of Cu-Zn superoxide dismutase overexpression of life span and resistance to oxidative stress in transgenic Drosophila melanogaster. Arch. Biochem. Biophys. 1993, 301, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhou, L.; Wang, Z.; Roberts, L.J.; Lin, X.; Zhao, Y.; Guo, Z. Overexpression of antioxidant enzymes in ApoE-deficient mice suppresses Benzo(a)pyrene-accelerated atherosclerosis. Atherosclerosis 2009, 207, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, H.; Ramesh, A.; Roberts, L.J.; Zhou, L.; Lin, X.; Zhao, Y.; Guo, Z. Overexpression of Cu/Zn-superoxide dismutase and/or catalase accelerates Benzo(a)pyrene detoxification by upregulation of the aryl hydrocarbon receptor in mouse endothelial cells. Free Radic. Biol. Med. 2009, 47, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Mockett, R.J.; Orr, W.C.; Rahmandar, J.J.; Benes, J.J.; Radyuk, S.N.; Klichko, V.I.; Sohal, R.S. Overexpression of Mn-containing superoxide dismutase in transgenic Drosophila melanogaster. Arch. Biochem. Biophys. 1999, 371, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, Z.N.; Anscher, M.S.; Folz, R.J.; Archer, E.; Huang, H.; Chen, L.; Golson, M.L.; Samulski, T.S.; Dewhirst, M.W.; Vujaskovic, Z. Overexpression of extracellular superoxide dismutase reduces acute radiation induced lung toxicity. BMC Cancer 2005, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Tower, J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol. Cell. Biol. 1999, 19, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Chen, Y.; Epstein, P.N. Suppression of doxorubicin cardiotoxicity by overexpression of catalase in the heart of transgenic mice. J. Biol. Chem. 1996, 271, 12610–12616. [Google Scholar] [CrossRef] [PubMed]
- Parihar, V.K.; Allen, B.D.; Tran, K.K.; Chmielewski, N.N.; Craver, B.M.; Martirosian, V.; Morganti, J.M.; Rosi, S.; Vlkolinsky, R.; Acharya, M.M.; et al. Targeted Overexpression of Mitochondrial Catalase Prevents Radiation-Induced Cognitive Dysfunction. Antioxid. Redox Signal. 2015, 22, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Liao, A.C.; Craver, B.M.; Tseng, B.P.; Tran, K.K.; Parihar, V.K.; Acharya, M.M.; Limoli, C.L. Mitochondrial-Targeted Human Catalase Affords Neuroprotection from Proton Irradiation. Radiat. Res. 2013, 180, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Moskalev, A.; Plyusnina, E.; Shaposhnikov, M.; Shilova, L.; Kazachenok, A.; Zhavoronkov, A. The role of D-GADD45 in oxidative, thermal and genotoxic stress resistance. Cell Cycle 2012, 11, 4222–4241. [Google Scholar] [CrossRef] [PubMed]
- Domen, J.; Gandy, K.L.; Weissman, I.L. Systemic overexpression of BCL-2 in the hematopoietic system protects transgenic mice from the consequences of lethal irradiation. Blood 1998, 91, 2272–2282. [Google Scholar] [PubMed]
- Dobrovol’skaya, E.V.; Solovyov, I.A.; Proshkina, E.N.; Moskalev, A.A. Effects of genes overactivation of circadian rhythms in different tissues to stress resistance and longevity of Drosophila melanogaster. Theor. Appl. Ecol. 2016, 32–40. [Google Scholar]
- Alcendor, R.R.; Gao, S.; Zhai, P.; Zablocki, D.; Holle, E.; Yu, X.; Tian, B.; Wagner, T.; Vatner, S.F.; Sadoshima, J. Sirt1 Regulates Aging and Resistance to Oxidative Stress in the Heart. Circ. Res. 2007, 100, 1512–1521. [Google Scholar] [CrossRef] [PubMed]
- Hwangbo, D.S.; Gershman, B.; Gersham, B.; Tu, M.-P.; Palmer, M.; Tatar, M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature 2004, 429, 562–566. [Google Scholar] [CrossRef] [PubMed]


| Gene (Gene ID *; Origin If Different) | Cells | Agents | R * | References |
|---|---|---|---|---|
| Genes involved in DNA damage recognition and repair | ||||
| RPA3 (6119) | Human nasopharyngeal carcinoma (CNE2, HK1) | X-ray | ↑ | [75] |
| XPA (7507) | SV-40 transformed primary human cells | UV | ↑ | [76] |
| APN1 (853746; yeast) coding homolog of mammalian APE1 | Chinese hamster (CHO-9) | MMS | ↑ | [77] |
| H2O2 | ↑ | [77] | ||
| APE1 (328) | Chinese hamster (CHO) | dioxolane cytidine | ↑ | [67] |
| Mammalian cells | γ-ray | 0 | [67,78] | |
| alkylating agents | 0 | [67,68,78] | ||
| Chinese hamster (CHO) | H2O2 | 0 | [67] | |
| mitomycin C, porfiromycin, daunorubicin and aziridinyl benzoquinone (drugs that are activated by reduction) | ↓ | [68] | ||
| Chinese hamster XRCC1-deficient (CHO) | alkylating agents | ↓ | [79] | |
| Chimeric MGMT (4255) + APE1 (328) | Human cervix adenocarcinoma (HeLa) | alkylating agents | ↑ | [80] |
| Ku70 (2547) | Human renal carcinoma 786-O | γ-ray | ↑ | [81] |
| Ku70 (2547; human) + Ku80 (34930; human) | Rat cell lines Rat-1 and R708 | X-ray | ↓ | [82] |
| DNA-PK (5591) | Human promyelocytic leukemia HL60 | adriamycin | ↑ | [83] |
| Rad51 (5888) | Mammalian cells | γ-ray | ↑ | [84,85] |
| Chinese hamster (V79) | etoposide, hydroxyurea, thymidine | ↑ | [86] | |
| Mouse hybridoma cells | mitomycin C | ↑ | [85] | |
| Prpf19 (27339) | Human umbilical vein/vascular endothelium cells (HUVECs) | bleomycin, DL-buthionine-sulfoximine | ↑ | [11] |
| ALC1 (9557) | Human osteosarcoma U2OS cells | phleomycin | ↓ | [87] |
| Lig III (3980) | Human cervix adenocarcinoma (HeLa S3) | MNNG | ↑ | [88] |
| DNA pol β (5423) | Chinese hamster (CHO) | cisplatin, melphalan, mechlorethamine | ↑↓ | [57] |
| Mouse embryo fibroblast (MEF) | MMS | ↑0↓ | [60] | |
| Tag (947137; E. coli) coding methyladenine DNA glycosylase I | Chinese hamster (V79) | MMS, MNU, EMS | ↑ | [89,90] |
| MNU, ENU | 0 | [90] | ||
| Murine fibroblast (NIH3T3) and murine H1 melanoma cells (B78) | MNU, MNNG, DMS, temozolomlde | 0 | [91] | |
| AlkA (947371; E. coli) coding methyladenine DNA glycosylase II | Chinese hamster (V79 and Irs1) | DMS, EMS, MMS | ↑ | [92] |
| MPG (4350) | Chinese hamster (V79 and Irs1) | DMS, EMS, MMS | ↑ | [92] |
| Chinese hamster (CHO) | MMS | ↓ | [93] | |
| bis-chloroethylnitrosourea, melphalan | 0 | [94] | ||
| DMS, EMS, MMS | 0 | [95] | ||
| MMS, MNNG | ↓ | [66] | ||
| Mouse embryo fibroblast (MEF) | temozolomide | ↓ | [96,97] | |
| FPG (946765; E. coli) coding homolog of mammalian OGG1 | Chinese hamster (CHO and V79) | γ-ray | ↑ | [98] |
| Chinese hamster (CHO) | aziridine | ↑ | [99] | |
| dOGG1 (31806) | Drosophila S2 cells | paraquat, H2O2 | ↓ | [100] |
| S-nitroso-N-acetylpenicillamine | ↑ | [100] | ||
| OGG1 (4968; human) | Chinese hamster (AA8 and AS52) | potassium bromate or [R]-1-[(10-chloro-4-oxo-3-phenyl-4H-benzo[a]quinolizine-1-yl)-carbonyl]-2-pyrrolidinemethanol plus light | ↑ | [101] |
| ERCC1 (2067; human) | Chinese hamster (AA8) | melphalan, cisplatin | ↓ | [102] |
| UV | 0 | [102] | ||
| NTH (947122; E.coli) | Chinese hamster (XRS7) | γ-ray | 0 | [103] |
| H2O2 | ↑ | [103] | ||
| bleomycin | ↓ | [103] | ||
| Ogt (945853; E. coli) | Mammalian cells | alkylating agents | ↑ | [104,105,106] |
| Ada (946710; E. coli) and its truncated and modified versions | Mammalian cells | alkylating agents | ↑ | [104,105,106,107,108,109,110,111,112,113,114,115,116,117] |
| Chinese hamster lung fibroblasts | dibromoalkanes | ↓ | [105] | |
| Chinese hamster (V79) | MMS, HN2 | 0 | [114] | |
| Chinese hamster (CHO) | UV, ENU | 0 | [112] | |
| MGMT (4255) and its modified versions | Mammalian cells | alkylating agents | ↑ | [112,118,119,120,121,122,123,124,125] |
| Chinese hamster (CHO) | UV, ENU | 0 | [112] | |
| alkB (946708; E. coli) | Human cervix adenocarcinoma (HeLa) | MMS, DMS | ↑ | [126] |
| Genes involved in detoxification and efflux of free radicals and xenobiotics | ||||
| SOD1 (6647) | Human lymphoblastoid cells (TK6) | γ-ray | 0 | [127] |
| Human primary lung fibroblasts (HPLF) | γ-ray | ↑ | [128] | |
| Astrocytes of mice | xanthine oxidase with hypoxanthine, menadione | ↑ | [129] | |
| Brain neurons of mice | S-nitroso-N-acetylpenicillamine, spermine-NONOate, diethylamine-NONOate | ↑ | [130] | |
| H2O2 | 0 | [130] | ||
| menadione | ↓ | [130] | ||
| Normal human keratinocytes | UV | 0 | [131] | |
| Human glioma cells (U118-9) | γ-ray | ↑ | [132] | |
| SOD2 (6648) | Human lung adenocarcinoma | cisplatin | ↑ | [133] |
| Human cells | γ-ray | ↑ | [127,128,134,135] | |
| Human lymphoblastoid cells (TK6) | paraquat | ↑ | [127] | |
| Human hepatocellular carcinoma cells (HLE) | X-ray | ↑ | [136] | |
| Human gastric carcinoma cells | doxorubicin | ↑ | [37] | |
| ALDH3A1 (218) | Human adenocarcinoma cells (MCF7) | 4-hydroxyperoxycyclophosphamide, doxorubicin, etoposide, 5-fluorouracil, γ-ray, H2O2 | ↑ | [137] |
| CAT (847) | Normal human keratinocytes | UV | ↑ | [131] |
| Mouse aortic endothelial cells (MAECs) | benzo(a)pyrene | ↑ | [138] | |
| TRX (41737) | Drosophila S2 Cells | H2O2 | ↑ | [139] |
| MTII (17750) | Chinese hamster ovary cells (K1-2) | Cadmium chloride, MNU, MNNG | ↑ | [140] |
| γ-ray, bleomycin, MMS, N-hydroxyethyl-N-hloroethylnitrosourea | 0 | [140] | ||
| Mouse C127 | cisplatin, melphalan, chlorambucil | ↑ | [141] | |
| 5-fluorouracil, vincristine | 0 | [141] | ||
| Mouse β-cell | streptozotocin | ↑ | [129] | |
| MTI (17748) | Mouse embryo fibroblasts (NIH/3T3) | tert-butyl hydroperoxide | ↑ | [142] |
| Chinese hamster (V79) | Amsacrine, menadione, arsenite, TPA | ↑ | [143] | |
| Zn(II) | ↑ | [144] | ||
| alkylating agents | 0 | [144] | ||
| Genes involved in control of proliferation and cell cycle | ||||
| CCND1 (595) | Human adenocarcinoma cells (MCF7) | γ-ray | ↓ | [145] |
| p21 (1026) | Glioma cells (T-98G, U-251MG with mutant p53 allele and U-87MG with wild-type p53). Medulloblastoma cells MED-3. | γ-ray | ↑ | [146] |
| Genes involved in regulation of apoptosis | ||||
| BCL2 (596) | Mice thymocytes | Ionizing radiation (not specified) | ↑ | [147] |
| Rat 6 fibroblast (R6) | UV | ↑ | [148] | |
| Human bladder cancer cells BIU87 | adriamycin | ↑ | [149] | |
| Mouse embryo fibroblasts (NIH/3T3) | γ-ray | ↑ | [150] | |
| Human breast cancer cells (MDA-MB-231) | γ-ray | ↑ | [150] | |
| Human non-small cell lung carcinoma (H1299) | Ionizing radiation (not specified) | ↓ | [151] | |
| Genes with other function | ||||
| USP22 (23326) | Human lung carcinoma cells (A549) | cisplatin | ↑ | [152] |
| IGF1R (3480) | Mammalian cells | γ-ray | ↑ | [153,154,155,156,157] |
| Sirt1 (23411) | Hepatocellular carcinoma cells (SK-Hep1) | doxorubicin | ↑ | [158] |
| Human skin fibroblasts (HS27) | UV | ↑ | [159] | |
| Human endometrial carcinoma cells (HHUA) | cisplatin | ↑ | [160] | |
| Human gastric cancer cells (SGC7901) | adriamycin, cisplatin, fluorouracil | ↑ | [161] | |
| Normal human foreskin fibroblasts (HCA2) | Endonuclease induced DBS | 0 | [162] | |
| Sirt2 (22933) | Normal human foreskin fibroblasts (HCA2) | Endonuclease induced DBS | 0 | [162] |
| NAMPT (10135) | Human prostate adenocarcinoma cells (LNCaP) | H2O2 | ↑ | [163] |
| VASH1 (22846) | Human umbilical vein/vascular endothelium cells (HUVECs) | H2O2 | ↑ | [164] |
| Sirt6 (51548) | Normal human foreskin fibroblasts (HCA2) | Endonuclease induced DBS, paraquat, neocarzinostatin | ↑ | [162] |
| Sirt7 (51547) | Mouse embryo fibroblasts (NIH/3T3) | doxorubicin | ↑ | [165] |
| Normal human foreskin fibroblasts (HCA2) | Endonuclease induced DBS | ↑ | [162] | |
| BRCC3 (79184) | Nasopharyngeal carcinoma cells (CNE2) | X-ray | ↑ | [166] |
| Bmi1 (12151) | Mice hematopoietic stem cells | γ-ray | 0 | [167] |
| STAT1 (6772) | Human head and neck squamous cell carcinoma cells (SCC-61) | X-ray | ↑ | [45] |
| SLC25A11 (67863) | Mouse motoneuron-like cells (NSC34) | H2O2, ethacrynic acid, sodium nitroprusside | ↑ | [168] |
| ICAM-3 (3385) | Human lung carcinoma cells (H1299) | γ-ray | ↑ | [40] |
| AKR1C3 (8644) | Human prostate cells (DU145) | 6 MV photons | ↑ | [169] |
| Pin1 (5300) | Cervix epidermoid carcinoma (Me180) | cisplatin | ↑ | [170] |
| PVT1 (5820) | Human cancer cell lines | cisplatin | ↑ | [171,172] |
| WRAP53 (55135) | Human osteosarcoma cells (U2OS) | γ-ray | ↑ | [173] |
| TRF2 (7014) | Human fibroblasts (MRC-5) | H2O2 | ↑ | [174] |
| Normal human foreskin fibroblasts (HCA2) | Endonuclease induced DBS | ↑ | [175] | |
| MYC (4609) | Normal human foreskin fibroblasts | γ-ray | ↓ | [176] |
| TEIF (57410) | Human cervix adenocarcinoma (HeLa) | H2O2 | ↑ | [177] |
| PARP1 (142) | Rat ovarian tumor cells (O-342) | γ-ray, MNNG | ↓ | [178] |
| cisplatin | 0 | [178] | ||
| Chinese hamster (C060) | γ-ray | ↓ | [179] | |
| Chinese hamster (CHO) | UV, MMS | ↑ | [180] | |
| HOTAIR (100124700) | Human ovarian carcinoma cells (2780) | cisplatin | ↑ | [181] |
| RPS3 (42761; Drosophila) | Human bone marrow cells from Fanconi anemia patients | mitomycin C | ↑ | [182] |
| Drosophila S2 cells | paraquat, H2O2 | ↓ | [100] | |
| S-nitroso-N-acetylpenicillamine | ↑ | [100] | ||
| RPS3 (6188) | Human skin fibroblasts | UV | ↑ | [183] |
| CAIII (54232; rat) | Mouse embryo fibroblasts (NIH/3T3) | H2O2 | ↑ | [184] |
| constitutively active PI3K p110 (170911) | Rat embryo fibroblasts (MR4) and human papilloma cells (RT4) | γ-ray | ↑ | [26] |
| p53 (7157) | Multidrug resistant human osteosarcoma cells (U-2OSR2 and KHOSR2) | taxol, cisplatin, doxorubicin | ↓ | [185] |
| Human non–small cell lung cancer (A549, H1299) and colon cancer cell lines (HCT116 p53+/+, HCT116 p53−/−) | bleomycin | ↓ | [186] | |
| Human non–small cell lung cancer (A549; H1299; H358) | cisplatin, paclitaxel | ↓ | [187] | |
| Human colon cancer cells (HT29) | γ-ray | ↓ 0 | [188] | |
| SMAR1 (54971) | Human adenocarcinoma cells (MCF7) | Irradiation by 89SrCl2 | ↓ | [189] |
| Gene (Gene ID *; Origin, If Different) | Object | Overexpression Specificity | Agents | R * | References |
|---|---|---|---|---|---|
| Genes involved in DNA damage recognition and repair | |||||
| mus210 (36697) | D. melanogaster | ubiquitous | γ-ray | 0 | [190] |
| paraquat | ↓ | [191] | |||
| mei9 (31373) | D. melanogaster | ubiquitous | γ-ray | ↓ | [190] |
| paraquat | ♂—↑; ♀—0 | [191] | |||
| neurospecific | paraquat | ↓ | [191] | ||
| Rrp1 (33500) | D. melanogaster | ubiquitous | paraquat | ♂—↑; ♀—0 | [191] |
| γ-ray | ↓ | [190] | |||
| Ku80 (34930) | D. melanogaster | ubiquitous | γ-ray | 0 | [190] |
| paraquat | ♂—↑; ♀—0 | [191] | |||
| Brca2 (37916) | D. melanogaster | ubiquitous | γ-ray | 0 | [190] |
| spnB (41746) | D. melanogaster | ubiquitous | γ-ray | 0 | [190] |
| dPrp19 (37123) | D. melanogaster | ubiquitous | paraquat, cisplatin | ♀—↑ | [192] |
| Ada (946710; E. coli) and its truncated and modified versions | M. musculus | ubiquitous | dimethylnitrosamine, diethylnitrosamine | ↑ | [193] |
| hepatic | MNU, nitrosodimethylamine | ↑ | [194] | ||
| MGMT (4255) and its modified versions | M. musculus | bone marrow | alkylating agents | ↑ | [119,120,124,195] |
| ubiquitous but predominantly in the thymus | alkylating agents | ↑ | [53,55,56,196,197,198] | ||
| epidermal | alkylating agents | ↑ | [51,199] | ||
| lung | 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone | ↑ | [54] | ||
| Genes involved in detoxification and efflux of free radicals and xenobiotics | |||||
| Gclc (53581) | D. melanogaster | ubiquitous | paraquat | ↑ | [200] |
| SOD1 (6647) | D. melanogaster | motorneurons | paraquat | ↑ | [201] |
| γ-ray | ↑ | [201] | |||
| ubiquitous | paraquat | 0 | [202] | ||
| M. musculus | ubiquitous | benzo(a)pyrene | ↑ | [203,204] | |
| SOD2 (36878) | D. melanogaster | ubiquitous | 100% O2 | 0 | [205] |
| EC-SOD (6649) | M. musculus | alveolar type II and nonciliated distal bronchial epithelial cells | 4-MV photons | ↑ | [206] |
| CAT (847) | D. melanogaster | ubiquitous | H2O2 | ↑ | [207] |
| M. musculus | heart-specific | doxorubicin | ↑ | [208] | |
| ubiquitous | benzo(a)pyrene | ↑ | [203,204] | ||
| proton irradiation | ↑ | [209,210] | |||
| MTII (17750) | M. musculus | ubiquitous | streptozotocin | ↑ | [129] |
| Genes involved in control of proliferation and cell cycle | |||||
| Mnk (35288) | D. melanogaster | neurospecific | paraquat | ↓ | [191] |
| dGADD45 (35646) | D. melanogaster | ubiquitous | γ-ray | ↓ | [190] |
| neurospecific | paraquat | ♂—↑; ♀—0 | [211] | ||
| γ-ray | 0 | [211] | |||
| Genes involved in regulation of apoptosis | |||||
| BCL2 (596; human) | M. musculus | ubiquitous | X-ray | ↑ | [212] |
| Genes with other function | |||||
| WRNexo (42208) | D. melanogaster | neurospecific | paraquat | ↓ | [191] |
| ubiquitous | γ-ray | 0 | [190] | ||
| Per (31251) | D. melanogaster | neurospecific | paraquat | ↑ | [213] |
| CLOCK (38872) | D. melanogaster | neurospecific | paraquat | ↑ | [213] |
| Cyc (40162) | D. melanogaster | neurospecific | paraquat | ↓ | [213] |
| IGF1R_(3480; human) | KSN nude M. musculus | tumor generated by transgenic HeLa cells | X-ray | ↑ | [153] |
| Sirt1 (93759) | M. musculus | heart-specific | paraquat | ↑ | [214] |
| VASH1 (22846; human) | M. musculus | intratracheally infected with adenovirus vector encoding human VASH1 | paraquat | ↑ | [164] |
| dFOXO (41709) | D. melanogaster | pericerebral fat body | paraquat | ↑ | [215] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velegzhaninov, I.O.; Ievlev, V.A.; Pylina, Y.I.; Shadrin, D.M.; Vakhrusheva, O.M. Programming of Cell Resistance to Genotoxic and Oxidative Stress. Biomedicines 2018, 6, 5. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines6010005
Velegzhaninov IO, Ievlev VA, Pylina YI, Shadrin DM, Vakhrusheva OM. Programming of Cell Resistance to Genotoxic and Oxidative Stress. Biomedicines. 2018; 6(1):5. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines6010005
Chicago/Turabian StyleVelegzhaninov, Ilya O., Vitaly A. Ievlev, Yana I. Pylina, Dmitry M. Shadrin, and Olesya M. Vakhrusheva. 2018. "Programming of Cell Resistance to Genotoxic and Oxidative Stress" Biomedicines 6, no. 1: 5. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines6010005