Mucosa-Associated Lymphoid Tissue 1 Is an Oncogene Inducing Cell Proliferation, Invasion, and Tumor Growth via the Upregulation of NF-κB Activity in Human Prostate Carcinoma Cells
Abstract
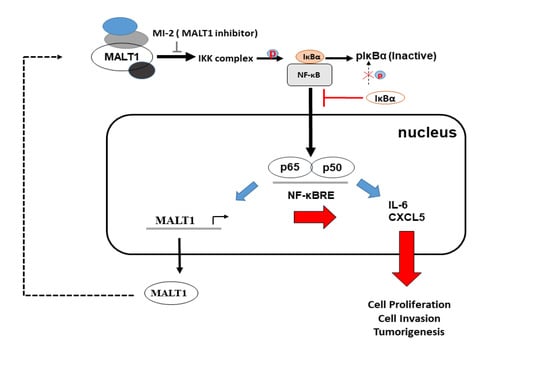
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. CyQUANT Cell Proliferation Assay
2.3. EdU Flow Cytometry Assay
2.4. Expression Vector Constructs and Stable Transfection
2.5. MALT1 CRISPR/dCas9 Lentiviral Activation Particles Transduction
2.6. Gene Knockdown
2.7. Nuclear and Cytoplasmic Extraction Assay
2.8. Immunoblot Assays
2.9. Real-Time Reverse Transcription-Polymerase Chain Reaction
2.10. Reporter Vector Constructs
2.11. Transient Transfection and Reporter Assay
2.12. Enzyme-Linked Immunosorbent Assay
2.13. Xenograft Animal Model
2.14. Soft Agar Cloning Assay
2.15. Matrigel Invasion Assay
2.16. Immunohistochemical Assay
2.17. NF-κB (p65) Transcription Factor Binding Assay
2.18. Statistical Analysis
3. Results
3.1. Expression of MALT1 in Prostate Carcinoma Cells and Prostate Tissues
3.2. Ectopic MALT1 Overexpression Induces IL-6 and CXCL5 via Activation of NF-κB Signaling to Enhance Cell Proliferation and Invasion In Vitro
3.3. Upregulation of Endogenous MALT1 Expression Activates NF-κB to Enhance Tumorigenesis In Vitro
3.4. Knockdown of MALT1 Attenuates Cell Proliferation and Cell Invasion In Vitro and Decreases Tumor Growth In Vivo
3.5. MALT1 Modulates NF-κB Activation in PC-3 Cells
3.6. NF-κB Activity Modulates MALT1 Expression in PC-3 Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Health Promotion Administration (HPA). Taiwan Cancer Registry. 2017. Available online: https://www.hpa.gov.tw/Pages/Detail.aspx?nodeid=269&pid=12235 (accessed on 16 March 2020).
- Scher, H.I.; Morrism, M.J.; Basch, E.; Heller, G. End points and outcomes in castration-resistant prostate cancer: From clinical trials to clinical practice. J. Clin. Oncol. 2011, 29, 3695–3704. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Bai, L.; Chen, W.; Xu, S. The NF-kB activation pathways, emerging molecular target for cancer prevention and therapy. Expert. Opin. Ther. Targets 2010, 14, 45–55. [Google Scholar] [CrossRef] [Green Version]
- Chaturvedi, M.M.; Sung, B.; Yadav, V.R.; Kannappan, R.; Aggarwal, B.B. NF-kB addiction and its role in cancer: ‘One size does not fit all’. Oncogene 2011, 30, 1615–1630. [Google Scholar] [CrossRef] [Green Version]
- Staal, J.; Beyaert, R. Inflammation and NF-kB signaling in prostate cancer: Mechanisms and clinical implications. Cells 2018, 7, 122. [Google Scholar] [CrossRef] [Green Version]
- Jin, R.; Yi, Y.; Yull, F.E.; Blackwell, T.S.; Clark, P.E.; Koyama, T.; Smith, J.A., Jr.; Matusik, R.J. NF-kB gene signature predicts prostate cancer progression. Mol. Cell. Pathobiol. 2014, 74, 2763–2772. [Google Scholar]
- Suh, J.; Payvandi, F.; Edelstein, L.C.; Amenta, P.S.; Zong, W.X.; Gelinas, C.; Rabson, A.B. Mechanisms of constitutive NF-kB activation in human prostate cancer cells. Prostate 2002, 52, 183–200. [Google Scholar] [CrossRef]
- Gasparian, A.V.; Yao, Y.J.; Kowalczyk, D.; Lyakh, L.A.; Karseladze, A.; Slaga, T.J.; Budunova, I.V. The role of IKK in constitutive activation of NF-kB transcription factor in prostate carcinoma cells. J. Cell Sci. 2002, 115, 141–151. [Google Scholar]
- Jin, R.J.; Lho, Y.; Connelly, L.; Wang, Y.; Yu, X.; Saint Jean, L.; Case, T.C.; Ellwood-Yen, K.; Sawyers, C.L.; Bhowmick, N.A.; et al. The nuclear factor-kB pathway controls the progression of prostate cancer to androgen-independent growth. Cancer Res. 2008, 68, 6762–6769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosset, A.-A.; Ouellet, V.; Caron, C. Validation of the prognostic value of NF-kB p65 in prostate cancer: A retrospective study using a large multi-institutional cohort of the Canadian Prostate Cancer Biomarker Network. PLoS Med. 2019, 16, e1002847. [Google Scholar] [CrossRef]
- Young, R.M.; Staudt, L.M. A new “brew” of MALT1 inhibitors. Cancer Cell 2012, 22, 706–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afonina, I.S.; Elton, L.; Carpentier, I.; Beyaert, R. MALT1—A universal soldier: Multiple strategies to ensure NF-kB activation and target gene expression. FEBS J. 2015, 282, 3286–3297. [Google Scholar] [CrossRef]
- Rosebeck, S.; Rehman, A.O.; Lucas, P.C.; McAllister-Lucas, L.M. From MALT lymphoma to the CBM signalosome: Three decades of discovery. Cell Cycle 2011, 10, 2485–2496. [Google Scholar] [CrossRef] [PubMed]
- Thome, M. Multifunctional roles for MALT1 in T-cell activation. Nat. Rev. Immunol. 2008, 8, 495–500. [Google Scholar] [CrossRef]
- Sun, L.; Deng, L.; Ea, C.K.; Xia, Z.P.; Chen, Z.J. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 2004, 14, 289–301. [Google Scholar] [CrossRef]
- Du, M.Q. MALT lymphoma: A paradigm of NF-kB dysregulation. Semin. Cancer Biol. 2016, 39, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Demeyer, A.; Staal, J.; Beyaert, R. Targeting MALT1 proteolytic activity in immunity, inflammation and disease: Good or bad? Trends Mol. Med. 2016, 22, 135–150. [Google Scholar] [CrossRef]
- Silva, L.C.; Ferreira-Strixino, J.; Fontana, L.C.; Rocha Gonsalves, A.M.; Serra, A.C.; Pineiro, M.; Canevari, R.A. Molecular analysis of apoptosis pathway after photodynamic therapy in breast cancer: Animal model study. Photodiagn. Photodyn. Ther. 2016, 14, 152–158. [Google Scholar] [CrossRef]
- Kingeter, L.M.; Schaefer, B.C. Malt1 and cIAP2-Malt1 as effectors of NF-kB activation: Kissing cousins or distant relatives? Cell Signal. 2010, 22, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Blonska, M.; Lin, X. CARMA1-mediated NF-kB and JNK activation in lymphocytes. Immunol. Rev. 2009, 228, 199–211. [Google Scholar] [CrossRef]
- Thome, M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat. Rev. Immunol. 2004, 4, 348–359. [Google Scholar] [CrossRef]
- McAllister-Lucas, L.M.; Baens, M.; Lucas, P.C. MALT1 protease: A new therapeutic target in B lymphoma and beyond? Clin. Cancer Res. 2011, 17, 6623–6631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekambaram, P.; Lee, J.L.; Hubel, N.E.; Hu, D.; Yerneni, S.; Campbell, P.G.; Pollock, N.; Klei, L.R.; Concel, V.J.; Delekta, P.C.; et al. The CARMA3-Bcl10-MALT1 signalosome drives NFkB activation and promotes aggressiveness in angiotensin II receptor-positive breast cancer. Cancer Res. 2018, 78, 1225–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, D.; Jiang, C.; Ma, Z.; Blonska, M.; You, M.J.; Lin, X. MALT1 is required for EGFR-induced NF-kB activation and contributes to EGFR-driven lung cancer progression. Oncogene 2016, 35, 919–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, G.; Jin, J.; Degan, S.; Tameze, Y.; Zhang, J.Y. MALT1 promotes melanoma progression through JNK/c-Jun signaling. Oncogenesis 2017, 6, e365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeh, C.N.; Chang, Y.C.; Su, Y.; Shin-Shian Hsu, D.; Cheng, C.T.; Wu, R.C.; Chung, Y.H.; Chiang, K.C.; Yeh, T.S.; Lu, M.L.; et al. Identification of MALT1 as both a prognostic factor and a potential therapeutic target of regorafenib in cholangiocarcinoma patients. Oncotarget 2017, 8, 113444–113459. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.S.; Tsui, K.H.; Lin, Y.H.; Hou, C.P.; Feng, T.H.; Juang, H.H. Migration and invasion enhancer 1 is an NF-kB-inducing gene enhancing the cell proliferation and invasion ability of human prostate carcinoma cells in vitro and in vivo. Cancers 2019, 11, 1486. [Google Scholar] [CrossRef] [Green Version]
- Chiang, K.C.; Tsui, K.H.; Lin, Y.H.; Hou, C.P.; Chang, K.S.; Tsai, H.H.; Shin, Y.S.; Chen, C.C.; Feng, T.H.; Juang, H.H. Antioxidation and antiapoptosis characteristics of heme oxygenase-1 enhance tumorigenesis of human prostate carcinoma cells. Transl. Oncol. 2020, 13, 102–112. [Google Scholar] [CrossRef]
- Lin, Y.H.; Tsui, K.H.; Chang, K.S.; Hou, C.P.; Feng, T.H.; Juang, H.H. Maspin is a PTEN-upregulated and p53-upregulated tumor suppressor gene and acts as an HDAC1 inhibitor in human bladder cancer. Cancers 2020, 12, 10. [Google Scholar] [CrossRef] [Green Version]
- Chiang, K.C.; Tsui, K.H.; Chung, L.C.; Yeh, C.N.; Feng, T.H.; Chen, W.T.; Chang, P.L.; Chiang, H.Y.; Juang, H.H. Cisplatin modulates B-cell translocation gene 2 to attenuate cell proliferation of prostate carcinoma cells in both p53-dependent and p53-independent pathways. Sci. Rep. 2014, 4, 5511. [Google Scholar] [CrossRef] [Green Version]
- Tsui, K.H.; Feng, T.H.; Hsieh, W.C.; Chang, P.L.; Juang, H.H. Expression of interleukin-6 is downregulated by 17-(allylamino)-17-demethoxygeldanamycin in human prostatic carcinoma cells. Acta Pharmacol. Sin. 2008, 29, 1334–1341. [Google Scholar] [CrossRef] [Green Version]
- Tsui, K.H.; Chang, Y.L.; Yang, P.S.; Hou, C.P.; Lin, Y.H.; Lin, B.W.; Feng, T.H.; Juang, H.H. The inhibitory effects of capillarisin on cell proliferation and invasion of prostate carcinoma cells. Cell Prolif. 2018, 51, e12429. [Google Scholar] [CrossRef] [Green Version]
- Tsui, K.H.; Chang, Y.L.; Feng, T.H.; Chung, L.C.; Lee, T.Y.; Chang, P.L.; Juang, H.H. Growth differentiation factor-15 upregulates interleukin-6 to promote tumorigenesis of prostate carcinoma PC-3 cells. J. Mol. Endocrinol. 2012, 49, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Nunes, J.J.; Pandey, S.K.; Yadav, A.; Goel, S.; Ateeq, B. Targeting NF-kB signaling by artesunate restores sensitivity of castrate-resistant prostate cancer cells to antiandrogens. Neoplasia 2017, 19, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Yamashita, H.Y.; Yu, S.; Wang, J.; Franco, O.E.; Wang, Y.; Hayward, S.W.; Matusik, R.J. Inhibition of NF-kB signaling restores responsiveness of castrate-resistant prostate cancer cells to anti-androgen treatment by decreasing androgen receptor-variant expression. Oncogene 2015, 34, 3700–3710. [Google Scholar] [CrossRef] [Green Version]
- Coornaert, B.; Baens, M.; Heyninck, K.; Bekaert, T.; Haegman, M.; Staal, J.; Sun, L.; Chen, Z.J.; Marynen, P.; Beyaert, R. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kB inhibitor A20. Nat. Immunol. 2008, 9, 263–271. [Google Scholar] [CrossRef]
- Hailfinger, S.; Nogai, H.; Pelzer, C.; Jaworski, M.; Cabalzar, K.; Charton, J.E.; Guzzardi, M.; Decaillet, C.; Grau, M.; Dorken, B.; et al. Malt1-dependent RelB cleavage promotes canonical NF-kB activation in lymphocytes and lymphoma cell lines. Proc. Natl. Acad. Sci. USA 2011, 108, 14596–14601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staal, J.; Driege, Y.; Bekaert, T.; Demeyer, A.; Muyllaert, D.; Van Damme, P.; Gevaert, K.; Beyaert, R. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 2011, 30, 1742–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.O.; Lou, W.; Hou, M.; de Miguel, F.; Gerber, L.; Gao, A.C. Interleukin-6 promotes androgen-independent growth in LNCaP human prostate cancer cells. Clin. Cancer Res. 2003, 9, 370–376. [Google Scholar] [PubMed]
- Kuo, P.L.; Chen, Y.H.; Chen, T.C.; Shen, K.H.; Hsu, Y.L. CXCL5/ENA78 increased cell migration and epithelial-to-mesenchymal transition of hormone-independent prostate cancer by early growth response-1/snail signaling pathway. J. Cell. Physiol. 2011, 226, 1224–1231. [Google Scholar] [CrossRef]
- Kumari, N.; Dwarakanath, B.S.; Das, A.; Bhatt, A.N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016, 37, 11553–11572. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chang, J.T.; Ho, Y.F.; Shyu, A.B. MiR-26 down-regulates TNF-alpha/NF-kB signalling and IL-6 expression by silencing HMGA1 and MALT1. Nucleic Acids Res. 2016, 44, 3772–3787. [Google Scholar] [CrossRef] [Green Version]
- Fontan, L.; Yang, C.; Kabaleeswaran, V.; Volpon, L.; Osborne, M.J.; Beltran, D.; Garcia, M.; Cerchietti, L.; Shaknovich, R.; Yang, S.N.; et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell 2012, 22, 812–824. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, K.; Singh, S.; Burke, T.R., Jr.; Grunberger, D. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-kB. Proc. Natl. Acad. Sci. USA 1996, 93, 9090–9095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uehata, T.; Iwassaki, H.; Vandenbon, A.; Matsushita, K.; Hemandez-Cuellar, E.; Kuniyoshi, K.; Satoch, T.; Mino, T.; Suzuki, Y.; Standley, D.M.; et al. Malt1-induced cleavage of regnase-1 in CD4+ helper T cells regulates immune activation. Cell 2013, 153, 1036–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmore, T.G. Introduction to NF-kB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef] [Green Version]
- Lessard, L.; Mes-Masson, A.M.; Lamarre, L.; Wall, L.; Lattouf, J.B.; Saad, F. NF-kB nuclear localization and its prognositic signifiance in prostate cancer. BJU Int. 2003, 91, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Holden, N.S.; Squires, P.E.; Kaur, M.; Bland, R.; Jones, C.E.; Newton, R. Phorbol ester-stimulated NF-kB-dependent transcription: Roles for isoforms of novel protein kinase C. Cell Signal. 2008, 20, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Catley, M.C.; Cambridge, L.M.; Nasuhara, Y.; Ito, K.; Chivers, J.E.; Beaton, A.; Holden, N.S.; Bergmann, M.W.; Barnes, P.J.; Newton, R. Inhibitors of protein kinase C (PKC) prevent activated transcription: Role of events downstream of NF-kB DNA binding. J. Biol. Chem. 2004, 279, 18457–18466. [Google Scholar] [CrossRef] [Green Version]
- Kucharzewska, P.; Maracle, C.X.; Jeucken, K.C.M.; van Hamburg, J.P.; Israelsson, E.; Furber, M.; Tas, S.W.; Olsson, H.K. NIK-IKK complex interaction controls NF-kB-dependent inflammatory activation of endothelium in response to LTβR ligation. J. Cell Sci. 2019, 132, jcs225615. [Google Scholar] [CrossRef] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsui, K.-H.; Chang, K.-S.; Sung, H.-C.; Hsu, S.-Y.; Lin, Y.-H.; Hou, C.-P.; Yang, P.-S.; Chen, C.-L.; Feng, T.-H.; Juang, H.-H. Mucosa-Associated Lymphoid Tissue 1 Is an Oncogene Inducing Cell Proliferation, Invasion, and Tumor Growth via the Upregulation of NF-κB Activity in Human Prostate Carcinoma Cells. Biomedicines 2021, 9, 250. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9030250
Tsui K-H, Chang K-S, Sung H-C, Hsu S-Y, Lin Y-H, Hou C-P, Yang P-S, Chen C-L, Feng T-H, Juang H-H. Mucosa-Associated Lymphoid Tissue 1 Is an Oncogene Inducing Cell Proliferation, Invasion, and Tumor Growth via the Upregulation of NF-κB Activity in Human Prostate Carcinoma Cells. Biomedicines. 2021; 9(3):250. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9030250
Chicago/Turabian StyleTsui, Ke-Hung, Kang-Shuo Chang, Hsin-Ching Sung, Shu-Yuan Hsu, Yu-Hsiang Lin, Chen-Pang Hou, Pei-Shan Yang, Chien-Lun Chen, Tsui-Hsia Feng, and Horng-Heng Juang. 2021. "Mucosa-Associated Lymphoid Tissue 1 Is an Oncogene Inducing Cell Proliferation, Invasion, and Tumor Growth via the Upregulation of NF-κB Activity in Human Prostate Carcinoma Cells" Biomedicines 9, no. 3: 250. https://0-doi-org.brum.beds.ac.uk/10.3390/biomedicines9030250