Genetic Testing for Neonatal Respiratory Disease
Abstract
:1. Introduction
1.1. Genetic Causes of Diffuse Lung Disease/Respiratory Distress Syndrome
1.2. Genetic Causes of Altered Lung Development and Pulmonary Hypertension
1.3. Genetic Causes of Transient Neonatal Respiratory Distress
1.4. Other Genes Responsible for Neonatal Lung Disease
1.5. Practical Issues of Genetic Testing for Neonatal Lung Disease
1.6. Candidates for Genetic Testing
2. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitsett, J.A.; Ohning, B.L.; Ross, G.; Meuth, J.; Weaver, T.; Holm, B.A.; Shapiro, D.L.; Notter, R.H. Hydrophobic surfactant-associated protein in whole lung surfactant and its importance for biophysical activity in lung surfactant extracts used for replacement therapy. Pediatr. Res. 1986, 20, 460–467. [Google Scholar] [CrossRef] [Green Version]
- Nogee, L.M. Genetic causes of surfactant protein abnormalities. Curr. Opin. Pediatr. 2019, 31, 330–339. [Google Scholar] [CrossRef]
- Suzuki, T.; Maranda, B.; Sakagami, T.; Catellier, P.; Couture, C.Y.; Carey, B.C.; Chalk, C.; Trapnell, B.C. Hereditary pulmonary alveolar proteinosis caused by recessive CSF2RB mutations. Eur. Respir. J. 2011, 37, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Sakagami, T.; Young, L.R.; Carey, B.C.; Wood, R.E.; Luisetti, M.; Wert, S.E.; Rubin, B.K.; Kevill, K.; Chalk, C.; et al. Hereditary pulmonary alveolar proteinosis: Pathogenesis, presentation, diagnosis, and therapy. Am. J. Respir. Crit. Care Med. 2010, 182, 1292–1304. [Google Scholar] [CrossRef] [Green Version]
- Nogee, L.M.; de Mello, D.E.; Dehner, L.P.; Colten, H.R. Brief report: Deficiency of pulmonary surfactant protein B in congenital alveolar proteinosis. N. Engl. J. Med. 1993, 328, 406–410. [Google Scholar] [CrossRef]
- Nogee, L.M.; Wert, S.E.; Proffit, S.A.; Hull, W.M.; Whitsett, J.A. Allelic heterogeneity in hereditary surfactant protein B (SP-B) deficiency. Am. J. Respir. Crit. Care Med. 2000, 161, 973–981. [Google Scholar] [CrossRef] [Green Version]
- Cameron, H.S.; Somaschini, M.; Carrera, P.; Hamvas, A.; Whitsett, J.A.; Wert, S.E.; Deutsch, G.; Nogee, L.M. A common mutation in the surfactant protein C gene associated with lung disease. J. Pediatr. 2005, 146, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Kroner, C.; Reu, S.; Teusch, V.; Schams, A.; Grimmelt, A.C.; Barker, M.; Brand, J.; Gappa, M.; Kitz, R.; Kramer, B.W.; et al. Genotype alone does not predict the clinical course of SFTPC deficiency in paediatric patients. Eur. Respir. J. 2015, 46, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Nogee, L.M.; Dunbar, A.E., 3rd; Wert, S.E.; Askin, F.; Hamvas, A.; Whitsett, J.A. A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N. Engl. J. Med. 2001, 344, 573–579. [Google Scholar] [CrossRef]
- Thomas, A.Q.; Lane, K.; Phillips, J., 3rd; Prince, M.; Markin, C.; Speer, M.; Schwartz, D.A.; Gaddipati, R.; Marney, A.; Johnson, J.; et al. Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am. J. Respir. Crit. Care Med. 2002, 165, 1322–1328. [Google Scholar] [CrossRef] [PubMed]
- Shulenin, S.; Nogee, L.M.; Annilo, T.; Wert, S.E.; Whitsett, J.A.; Dean, M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N. Engl. J. Med. 2004, 350, 1296–1303. [Google Scholar] [CrossRef] [Green Version]
- Wambach, J.A.; Casey, A.M.; Fishman, M.P.; Wegner, D.J.; Wert, S.E.; Cole, F.S.; Hamvas, A.; Nogee, L.M. Genotype-phenotype correlations for infants and children with ABCA3 deficiency. Am. J. Respir. Crit. Care Med. 2014, 189, 1538–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullard, J.E.; Wert, S.E.; Nogee, L.M. ABCA3 deficiency: Neonatal respiratory failure and interstitial lung disease. Semin. Perinatol. 2006, 30, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Kroner, C.; Wittmann, T.; Reu, S.; Teusch, V.; Klemme, M.; Rauch, D.; Hengst, M.; Kappler, M.; Cobanoglu, N.; Sismanlar, T.; et al. Lung disease caused by ABCA3 mutations. Thorax 2017, 72, 213–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullard, J.E.; Wert, S.E.; Whitsett, J.A.; Dean, M.; Nogee, L.M. ABCA3 mutations associated with pediatric interstitial lung disease. Am. J. Respir. Crit. Care Med. 2005, 172, 1026–1031. [Google Scholar] [CrossRef]
- Wambach, J.A.; Yang, P.; Wegner, D.J.; Heins, H.B.; Kaliberova, L.N.; Kaliberov, S.A.; Curiel, D.T.; White, F.V.; Hamvas, A.; Hackett, B.P.; et al. Functional Characterization of ATP-Binding Cassette Transporter A3 Mutations from Infants with Respiratory Distress Syndrome. Am. J. Respir. Cell Mol. Biol. 2016, 55, 716–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wambach, J.A.; Yang, P.; Wegner, D.J.; Heins, H.B.; Luke, C.; Li, F.; White, F.V.; Cole, F.S. Functional Genomics of ABCA3 Variants. Am. J. Respir. Cell Mol. Biol. 2020, 63, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Wambach, J.A.; Wegner, D.J.; Depass, K.; Heins, H.; Druley, T.E.; Mitra, R.D.; An, P.; Zhang, Q.; Nogee, L.M.; Cole, F.S.; et al. Single ABCA3 mutations increase risk for neonatal respiratory distress syndrome. Pediatrics 2012, 130, e1575–e1582. [Google Scholar] [CrossRef] [Green Version]
- Legendre, M.; Butt, A.; Borie, R.; Debray, M.P.; Bouvry, D.; Filhol-Blin, E.; Desroziers, T.; Nau, V.; Copin, B.; Dastot-Le Moal, F.; et al. Functional assessment and phenotypic heterogeneity of SFTPA1 and SFTPA2 mutations in interstitial lung diseases and lung cancer. Eur. Respir. J. 2020, 56. [Google Scholar] [CrossRef]
- van Moorsel, C.H.; Ten Klooster, L.; van Oosterhout, M.F.; de Jong, P.A.; Adams, H.; Wouter van Es, H.; Ruven, H.J.; van der Vis, J.J.; Grutters, J.C. SFTPA2 Mutations in Familial and Sporadic Idiopathic Interstitial Pneumonia. Am. J. Respir. Crit. Care Med. 2015, 192, 1249–1252. [Google Scholar] [CrossRef]
- Wang, Y.; Kuan, P.J.; Xing, C.; Cronkhite, J.T.; Torres, F.; Rosenblatt, R.L.; DiMaio, J.M.; Kinch, L.N.; Grishin, N.V.; Garcia, C.K. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am. J. Hum. Genet. 2009, 84, 52–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gower, W.A.; Nogee, L.M. Candidate gene analysis of the surfactant protein D gene in pediatric diffuse lung disease. J. Pediatr. 2013, 163, 1778–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamvas, A.; Deterding, R.R.; Wert, S.E.; White, F.V.; Dishop, M.K.; Alfano, D.N.; Halbower, A.C.; Planer, B.; Stephan, M.J.; Uchida, D.A.; et al. Heterogeneous pulmonary phenotypes associated with mutations in the thyroid transcription factor gene NKX2-1. Chest 2013, 144, 794–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krude, H.; Schutz, B.; Biebermann, H.; von Moers, A.; Schnabel, D.; Neitzel, H.; Tonnies, H.; Weise, D.; Lafferty, A.; Schwarz, S.; et al. Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2-1 haploinsufficiency. J. Clin. Investig. 2002, 109, 475–480. [Google Scholar] [CrossRef]
- Breedveld, G.J.; van Dongen, J.W.; Danesino, C.; Guala, A.; Percy, A.K.; Dure, L.S.; Harper, P.; Lazarou, L.P.; van der Linde, H.; Joosse, M.; et al. Mutations in TITF-1 are associated with benign hereditary chorea. Hum. Mol. Genet. 2002, 11, 971–979. [Google Scholar] [CrossRef] [Green Version]
- Galambos, C.; Levy, H.; Cannon, C.L.; Vargas, S.O.; Reid, L.M.; Cleveland, R.; Lindeman, R.; deMello, D.E.; Wert, S.E.; Whitsett, J.A.; et al. Pulmonary pathology in thyroid transcription factor-1 deficiency syndrome. Am. J. Respir. Crit. Care Med. 2010, 182, 549–554. [Google Scholar] [CrossRef] [Green Version]
- Galambos, C.; Sims-Lucas, S.; Ali, N.; Gien, J.; Dishop, M.K.; Abman, S.H. Intrapulmonary vascular shunt pathways in alveolar capillary dysplasia with misalignment of pulmonary veins. Thorax 2015, 70, 84–85. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.J.; Murali, C.; Pogoriler, J.; Frank, D.B.; Handler, S.S.; Deardorff, M.A.; Hopper, R.K. Histopathologic and Genetic Features of Alveolar Capillary Dysplasia with Atypical Late Presentation and Prolonged Survival. J. Pediatr. 2019, 210, 214–219.e2. [Google Scholar] [CrossRef]
- Towe, C.T.; White, F.V.; Grady, R.M.; Sweet, S.C.; Eghtesady, P.; Wegner, D.J.; Sen, P.; Szafranski, P.; Stankiewicz, P.; Hamvas, A.; et al. Infants with Atypical Presentations of Alveolar Capillary Dysplasia with Misalignment of the Pulmonary Veins Who Underwent Bilateral Lung Transplantation. J. Pediatr. 2018, 194, 158–164.e1. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Yang, Y.; Navarro, C.; Silva, I.; Szafranski, P.; Kolodziejska, K.E.; Dharmadhikari, A.V.; Mostafa, H.; Kozakewich, H.; Kearney, D.; et al. Novel FOXF1 mutations in sporadic and familial cases of alveolar capillary dysplasia with misaligned pulmonary veins imply a role for its DNA binding domain. Hum. Mutat. 2013, 34, 801–811. [Google Scholar] [CrossRef] [Green Version]
- Stankiewicz, P.; Sen, P.; Bhatt, S.S.; Storer, M.; Xia, Z.; Bejjani, B.A.; Ou, Z.; Wiszniewska, J.; Driscoll, D.J.; Maisenbacher, M.K.; et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am. J. Hum. Genet. 2009, 84, 780–791. [Google Scholar] [CrossRef] [Green Version]
- Szafranski, P.; Dharmadhikari, A.V.; Wambach, J.A.; Towe, C.T.; White, F.V.; Grady, R.M.; Eghtesady, P.; Cole, F.S.; Deutsch, G.; Sen, P.; et al. Two deletions overlapping a distant FOXF1 enhancer unravel the role of lncRNA LINC01081 in etiology of alveolar capillary dysplasia with misalignment of pulmonary veins. Am. J. Med. Genet. A 2014, 164A, 2013–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szafranski, P.; Gambin, T.; Dharmadhikari, A.V.; Akdemir, K.C.; Jhangiani, S.N.; Schuette, J.; Godiwala, N.; Yatsenko, S.A.; Sebastian, J.; Madan-Khetarpal, S.; et al. Pathogenetics of alveolar capillary dysplasia with misalignment of pulmonary veins. Hum. Genet. 2016, 135, 569–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szafranski, P.; Stankiewicz, P. Long Non-Coding RNA FENDRR: Gene Structure, Expression, and Biological Relevance. Genes 2021, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.; Karolak, J.A.; Deutsch, G.; Gambin, T.; Popek, E.; Isidor, B.; Szafranski, P.; Le Caignec, C.; Stankiewicz, P. Clinical, Histopathological, and Molecular Diagnostics in Lethal Lung Developmental Disorders. Am. J. Respir. Crit. Care Med. 2019, 200, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Szafranski, P.; Coban-Akdemir, Z.H.; Rupps, R.; Grazioli, S.; Wensley, D.; Jhangiani, S.N.; Popek, E.; Lee, A.F.; Lupski, J.R.; Boerkoel, C.F.; et al. Phenotypic expansion of TBX4 mutations to include acinar dysplasia of the lungs. Am. J. Med. Genet. A 2016, 170, 2440–2444. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Gonzaga-Jauregui, C.; Welch, C.L.; Ma, L.; Qi, H.; King, A.K.; Krishnan, U.; Rosenzweig, E.B.; Ivy, D.D.; Austin, E.D.; et al. Exome Sequencing in Children With Pulmonary Arterial Hypertension Demonstrates Differences Compared With Adults. Circ. Genom. Precis. Med. 2018, 11, e001887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karolak, J.A.; Vincent, M.; Deutsch, G.; Gambin, T.; Cogne, B.; Pichon, O.; Vetrini, F.; Mefford, H.C.; Dines, J.N.; Golden-Grant, K.; et al. Complex Compound Inheritance of Lethal Lung Developmental Disorders Due to Disruption of the TBX-FGF Pathway. Am. J. Hum. Genet. 2019, 104, 213–228. [Google Scholar] [CrossRef] [Green Version]
- Suhrie, K.; Pajor, N.M.; Ahlfeld, S.K.; Dawson, D.B.; Dufendach, K.R.; Kitzmiller, J.A.; Leino, D.; Lombardo, R.C.; Smolarek, T.A.; Rathbun, P.A.; et al. Neonatal Lung Disease Associated with TBX4 Mutations. J. Pediatr. 2019, 206, 286–292.e1. [Google Scholar] [CrossRef]
- Lopez-Andreu, J.A.; Hidalgo-Santos, A.D.; Fuentes-Castello, M.A.; Mancheño-Franch, N.; Cerón-Pérez, J.A.; Esteban-Ricós, M.J.; Pedrola-Vidal, L.; Nogee, L.M. Delayed Presentation and Prolonged Survival of a Child with Surfactant Protein B Deficiency. J. Pediatr. 2017, 190, 268–270.e1. [Google Scholar] [CrossRef]
- Davis, S.D.; Ferkol, T.W.; Rosenfeld, M.; Lee, H.S.; Dell, S.D.; Sagel, S.D.; Milla, C.; Zariwala, M.A.; Pittman, J.E.; Shapiro, A.J.; et al. Clinical features of childhood primary ciliary dyskinesia by genotype and ultrastructural phenotype. Am. J. Respir. Crit. Care Med. 2015, 191, 316–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holzmann, D.; Felix, H. Neonatal respiratory distress syndrome--A sign of primary ciliary dyskinesia? Eur. J. Pediatr. 2000, 159, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Leigh, M.W.; Ferkol, T.W.; Davis, S.D.; Lee, H.S.; Rosenfeld, M.; Dell, S.D.; Sagel, S.D.; Milla, C.; Olivier, K.N.; Sullivan, K.M.; et al. Clinical Features and Associated Likelihood of Primary Ciliary Dyskinesia in Children and Adolescents. Ann. Am. Thorac. Soc. 2016, 13, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.J.; Davis, S.D.; Polineni, D.; Manion, M.; Rosenfeld, M.; Dell, S.D.; Chilvers, M.A.; Ferkol, T.W.; Zariwala, M.A.; Sagel, S.D.; et al. Diagnosis of Primary Ciliary Dyskinesia. An Official American Thoracic Society Clinical Practice Guideline. Am. J. Respir. Crit Care Med. 2018, 197, e24–e39. [Google Scholar] [CrossRef] [PubMed]
- Mullowney, T.; Manson, D.; Kim, R.; Stephens, D.; Shah, V.; Dell, S. Primary ciliary dyskinesia and neonatal respiratory distress. Pediatrics 2014, 134, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goutaki, M.; Halbeisen, F.S.; Barbato, A.; Crowley, S.; Harris, A.; Hirst, R.A.; Karadag, B.; Martinu, V.; Morgan, L.; O’Callaghan, C.; et al. Late Diagnosis of Infants with PCD and Neonatal Respiratory Distress. J. Clin. Med. 2020, 9, 2871. [Google Scholar] [CrossRef]
- Davis, S.D.; Rosenfeld, M.; Lee, H.S.; Ferkol, T.W.; Sagel, S.D.; Dell, S.D.; Milla, C.; Pittman, J.E.; Shapiro, A.J.; Sullivan, K.M.; et al. Primary Ciliary Dyskinesia: Longitudinal Study of Lung Disease by Ultrastructure Defect and Genotype. Am. J. Respir.Crit. Care Med. 2019, 199, 190–198. [Google Scholar] [CrossRef]
- Horani, A.; Ferkol, T.W.; Dutcher, S.K.; Brody, S.L. Genetics and biology of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2016, 18, 18–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, M.; Kasper, B.; Bohring, A.; Rutsch, F.; Kluger, G.; Hoffjan, S.; Spranger, S.; Behnecke, A.; Ferbert, A.; Hahn, A.; et al. 47 patients with FLNA associated periventricular nodular heterotopia. Orphanet. J. Rare Dis. 2015, 10, 134. [Google Scholar] [CrossRef] [Green Version]
- Kinane, T.B.; Lin, A.E.; Lahoud-Rahme, M.; Westra, S.J.; Mark, E.J. Case 4-2017. A 2-Month-Old Girl with Growth Retardation and Respiratory Failure. N. Engl. J. Med. 2017, 376, 562–574. [Google Scholar] [CrossRef]
- Lord, A.; Shapiro, A.J.; Saint-Martin, C.; Claveau, M.; Melancon, S.; Wintermark, P. Filamin A mutation may be associated with diffuse lung disease mimicking bronchopulmonary dysplasia in premature newborns. Respir. Care 2014, 59, e171–e177. [Google Scholar] [CrossRef] [Green Version]
- Catteruccia, M.; Verrigni, D.; Martinelli, D.; Torraco, A.; Agovino, T.; Bonafe, L.; D’Amico, A.; Donati, M.A.; Adorisio, R.; Santorelli, F.M.; et al. Persistent pulmonary arterial hypertension in the newborn (PPHN): A frequent manifestation of TMEM70 defective patients. Mol. Genet. Metab. 2014, 111, 353–359. [Google Scholar] [CrossRef]
- Has, C.; Sparta, G.; Kiritsi, D.; Weibel, L.; Moeller, A.; Vega-Warner, V.; Waters, A.; He, Y.; Anikster, Y.; Esser, P.; et al. Integrin alpha3 mutations with kidney, lung, and skin disease. N. Engl. J. Med. 2012, 366, 1508–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Jesus, A.A.; Marrero, B.; Yang, D.; Ramsey, S.E.; Sanchez, G.A.M.; Tenbrock, K.; Wittkowski, H.; Jones, O.Y.; Kuehn, H.S.; et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 2014, 371, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Dimmock, D.P.; Clark, M.M.; Gaughran, M.; Cakici, J.A.; Caylor, S.A.; Clarke, C.; Feddock, M.; Chowdhury, S.; Salz, L.; Cheung, C.; et al. An RCT of Rapid Genomic Sequencing among Seriously Ill Infants Results in High Clinical Utility, Changes in Management, and Low Perceived Harm. Am. J. Hum. Genet. 2020, 107, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Farnaes, L.; Hildreth, A.; Sweeney, N.M.; Clark, M.M.; Chowdhury, S.; Nahas, S.; Cakici, J.A.; Benson, W.; Kaplan, R.H.; Kronick, R.; et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom. Med. 2018, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kingsmore, S.F. Is Rapid Exome Sequencing Standard of Care in the Neonatal and Pediatric Intensive Care Units? J. Pediatr. 2020. [Google Scholar] [CrossRef]
- Meng, L.; Pammi, M.; Saronwala, A.; Magoulas, P.; Ghazi, A.R.; Vetrini, F.; Zhang, J.; He, W.; Dharmadhikari, A.V.; Qu, C.; et al. Use of Exome Sequencing for Infants in Intensive Care Units: Ascertainment of Severe Single-Gene Disorders and Effect on Medical Management. JAMA Pediatr. 2017, 171, e173438. [Google Scholar] [CrossRef]
- Nogee, L.M.; Hamvas, A. The past and future of genetics in pulmonary disease: You can teach an old dog new tricks. Pediatr. Pulmonol. 2020, 55, 1789–1793. [Google Scholar] [CrossRef]
- Clark, M.M.; Hildreth, A.; Batalov, S.; Ding, Y.; Chowdhury, S.; Watkins, K.; Ellsworth, K.; Camp, B.; Kint, C.I.; Yacoubian, C.; et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef]
- Kingsmore, S.F.; Cakici, J.A.; Clark, M.M.; Gaughran, M.; Feddock, M.; Batalov, S.; Bainbridge, M.N.; Carroll, J.; Caylor, S.A.; Clarke, C.; et al. A Randomized, Controlled Trial of the Analytic and Diagnostic Performance of Singleton and Trio, Rapid Genome and Exome Sequencing in Ill Infants. Am. J. Hum. Genet. 2019, 105, 719–733. [Google Scholar] [CrossRef] [PubMed]
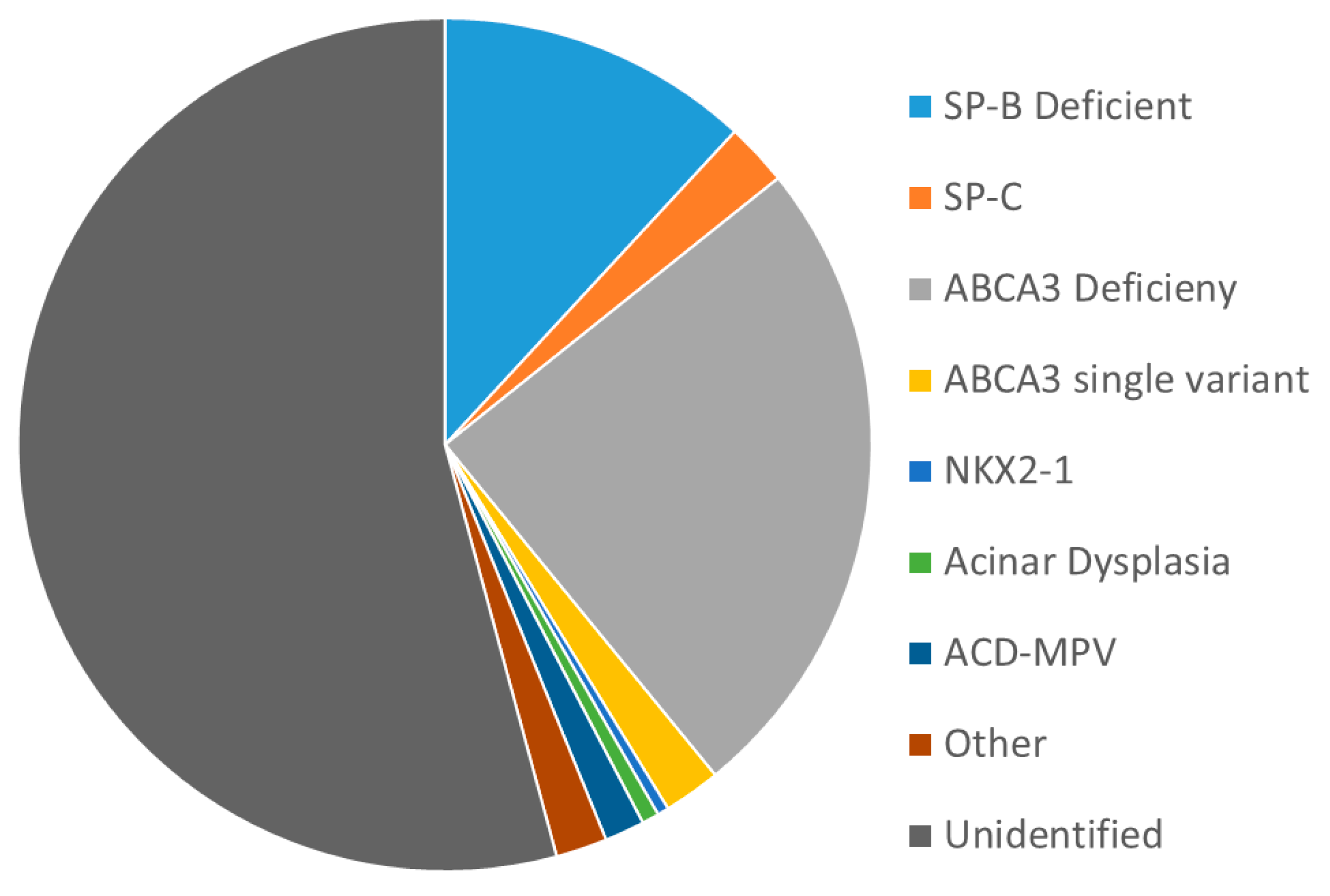
| Protein | SP-B | SP-C | ABCA3 | SP-A | SP-D | TTF−1 | GM-CSF Receptor [3,4] |
|---|---|---|---|---|---|---|---|
| Gene | SFTPB | SFTPC | ABCA3 | SFTPA1 SFTPA2 | SFTPD | NKX2–1 | CSFR2A CSFR2B |
| Pulmonary Phenotypes | RDS | ILD PF RDS | RDS PPHN ILD PF | PF Lung cancer | None yet known | RDS ILD Recurrent Infection | Alveolar Proteinosis |
| Inheritance | AR | AD sporadic | AR | AD sporadic | N.A. | Sporadic AD | AR |
| Prognosis | Rapidly fatal | Variable | ~60% rapidly fatal; ~40% variable | Generally adult onset, progressive | N.A. | Variable | Childhood to adult onset; variable |
| Incidence | <1 in 1,000,000 | Unknown | Uncertain, 1 in 10 K to 1 in 20 K | Unknown | N.A. | Unknown | Unknown |
| Category | Clinical Phenotype(s) | Genes | Extrapulmonary |
|---|---|---|---|
| Diffuse Lung Disease/ Surfactant Dysfunction | RDS PPHN | ABCA3 SFTPB SFTPC NKX2–1 | Isolated Lung Disease Hypothyroidism and neurological abnormalities |
| Pulmonary Hypertension Pulmonary Hypoplasia | PPHN | FOXF1 TBX4, FGF10 TMEM70 [52] | Multiple other organ systems Skeletal abnormalities PAH in older children Hypertrophic cardiomyopathy Hypotonia Hyperammonemia |
| Primary Ciliary Dyskinesia | Respiratory Distress Congenital Pneumonia “Wet” cough (in infancy) | Multiple | Situs Inversus Situs Ambiguous |
| Other | RDS Hyperinflation, “BPD” RDS | ITGA3 [53] FNLA (Filamin A) TMEM170 | Skin and Renal CNS heterotopias Cardiac and Skeletal Skin and joint, immune dysfunction, SAVI (“STING-associated vasculitis of infancy”) [54] |
| Clinical Phenotype | Factors Prompting Suspicion for Genetic Cause | Possible Genes | Additional Clinical Features |
|---|---|---|---|
| RDS, full-term | Lack of risk factors for RDS including: preterm gestation (<36 weeks) IDM, operative delivery without labor, clinical suspicion for infection. | SFTPB ABCA3 NKX2–1 SFTPC | NKX2–1 may be associated with hypothyroidism, neurological symptoms |
| RDS, preterm | Severity and/or persistence of disease out of proportion to that expected for infant’s gestational age or clinical history (SGA, likelihood of congenital pneumonia) | ABCA3 FLNA | Associated with monoallelic variants Hyperinflated CXR |
| PPHN | Lack of risk factors for PPHN (MAS, perinatal depression) or pulmonary hypoplasia (severe oligo or anhydramnios due to obstructive uropathy or early, prolonged ROM) | FOXF1 TBX4, FGF10 | Extrapulmonary manifestations common |
| Unexplained respiratory distress | Onset > 12 h Need for supplemental oxygen >48 h Lobar Infiltrates | PCD genes | ~50% with abnormal situs |
| Family history of neonatal lung disease, diffuse lung disease in older children or young adults, consanguinity should prompt early investigation | |||
| Method | Advantages | Disadvantages |
|---|---|---|
| Sanger Sequencing |
|
|
| NGS panels (e.g., “surfactant protein gene panel” or “congenital hypotonia gene panel”) |
|
|
| WES |
|
|
| WGS |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nogee, L.M.; Ryan, R.M. Genetic Testing for Neonatal Respiratory Disease. Children 2021, 8, 216. https://0-doi-org.brum.beds.ac.uk/10.3390/children8030216
Nogee LM, Ryan RM. Genetic Testing for Neonatal Respiratory Disease. Children. 2021; 8(3):216. https://0-doi-org.brum.beds.ac.uk/10.3390/children8030216
Chicago/Turabian StyleNogee, Lawrence M., and Rita M. Ryan. 2021. "Genetic Testing for Neonatal Respiratory Disease" Children 8, no. 3: 216. https://0-doi-org.brum.beds.ac.uk/10.3390/children8030216






