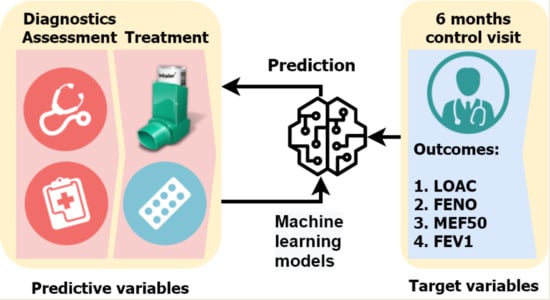
Predicting Treatment Outcomes Using Explainable Machine Learning in Children with Asthma
Abstract
:1. Introduction
Our Aim and Contribution
2. Materials and Methods
2.1. Population Studied
2.2. Response Variables
2.3. Data Preparation and Balancing
2.4. Machine Learning
3. Results
3.1. Performance in Prediction of Treatment Outcomes
3.2. Model Interpretation
3.3. The Need for Machine Learning
4. Discussion
5. Conclusions
- With respect to asthma chronicity, the assessment period of 6 months may not be enough for valid predictions. Longitudinal and prospective studies are essential;
- Additional studies involving larger numbers of patients with even more clinically relevant parameters are required to increase the success of the treatment outcome prediction and, further, the characterization of specific disease phenotypes and endotypes.
- With respect to asthma chronicity, the assessment period of 6 months may not be enough for valid predictions. Longitudinal and prospective studies are essential;
- Additional studies involving larger numbers of patients with even more clinically relevant parameters are required to increase the success of the treatment outcome prediction and the further characterization of specific disease phenotypes and endotypes;
- Although we tried to encompass both the objectively measured (lung function and FENO) and subjective target variables (asthma control), a consensus in the choice of the primary study endpoints is required.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamburg, M.A.; Collins, F.S. The Path to Personalized Medicine. N. Engl. J. Med. 2010, 363, 301–304. [Google Scholar] [CrossRef]
- Saglani, S.; Custovic, A. Childhood asthma: Advances using machine learning and mechanistic studies. Am. J. Respir. Crit. Care Med. 2019, 199, 414–422. [Google Scholar] [CrossRef]
- Pavord, I.D.; Beasley, R.; Agusti, A.; Anderson, G.P.; Bel, E.; Brusselle, G.; Cullinan, P.; Custovic, A.; Ducharme, F.M.; Fahy, J.V.; et al. After asthma: Redefining airways diseases. Lancet 2018, 391, 350–400. [Google Scholar] [CrossRef]
- Custovic, A.; Belgrave, D.; Lin, L.; Bakhsoliani, E.; Telcian, A.G.; Solari, R.; Murray, C.S.; Walton, R.P.; Curtin, J.; Edwards, M.R.; et al. Cytokine responses to rhinovirus and development of asthma, allergic sensitization, and respiratory infections during childhood. Am. J. Respir. Crit. Care Med. 2018, 197, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Blais, L.; Kettani, F.Z.; Lemire, C.; Beauchesne, M.F.; Perreault, S.; Elftouh, N.; Ducharme, F.M. Inhaled corticosteroids vs. leukotriene-receptor antagonists and asthma exacerbations in children. Respir. Med. 2011, 105, 846–855. [Google Scholar] [CrossRef] [Green Version]
- Turkalj, M.; Erceg, D. Terapijski pristup astmi u djece. Medicus 2013, 22, 49–56. [Google Scholar]
- Wu, W.; Bleecker, E.; Moore, W.; Busse, W.W.; Castro, M.; Chung, K.F.; Calhoun, W.J.; Erzurum, S.; Gaston, B.; Israel, E.; et al. Unsupervised phenotyping of Severe Asthma Research Program participants using expanded lung data. J. Allergy Clin. Immunol. 2014, 133, 1280–1288. [Google Scholar] [CrossRef] [Green Version]
- Szefler, S.J.; Phillips, B.R.; Martinez, F.D.; Chinchilli, V.M.; Lemanske, R.F.; Strunk, R.C.; Zeiger, R.S.; Larsen, G.; Spahn, J.D.; Bacharier, L.B.; et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J. Allergy Clin. Immunol. 2005, 115, 233–242. [Google Scholar] [CrossRef]
- Chung, K.F.; Adcock, I.M. Clinical phenotypes of asthma should link up with disease mechanisms. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Belgrave, D.; Cassidy, R.; Stamate, D.; Custovic, A.; Fleming, L.; Bush, A.; Saglani, S. Predictive modelling strategies to understand heterogeneous manifestations of asthma in early life. In Proceedings of the 16th IEEE International Conference on Machine Learning and Applications (ICMLA), Cancun, Mexico, 18–21 December 2017; Institute of Electrical and Electronics Engineers Inc.: Piscataway, NJ, USA, 2017; pp. 68–75. [Google Scholar]
- Ross, M.K.; Yoon, J.; Van Der Schaar, A.; Van Der Schaar, M. Discovering pediatric asthma phenotypes on the basis of response to controller medication using machine learning. Ann. Am. Thorac. Soc. 2018, 15, 49–58. [Google Scholar] [CrossRef]
- Luo, G.; Nkoy, F.L.; Stone, B.L.; Schmick, D.; Johnson, M.D. A systematic review of predictive models for asthma development in children Clinical decision-making, knowledge support systems, and theory. BMC Med. Inform. Decis. Mak. 2015, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoos, A.M.M.; Chawes, B.L.; Rasmussen, M.A.; Bloch, J.; Bønnelykke, K.; Bisgaard, H. Atopic endotype in childhood. J. Allergy Clin. Immunol. 2016, 137, 844–851.e4. [Google Scholar] [CrossRef] [Green Version]
- Bornelöv, S.; Sääf, A.; Melén, E.; Bergström, A.; Torabi Moghadam, B.; Pulkkinen, V.; Acevedo, N.; Orsmark Pietras, C.; Ege, M.; Braun-Fahrländer, C.; et al. Rule-Based Models of the Interplay between Genetic and Environmental Factors in Childhood Allergy. PLoS ONE 2013, 8, e80080. [Google Scholar] [CrossRef] [Green Version]
- Schneeberger, D.; Stöger, K.; Holzinger, A. The European Legal Framework for Medical AI. In Proceedings of the Lecture Notes in Computer Science; Springer: Berlin, Germany, 2020; Volume 12279, pp. 209–226. [Google Scholar]
- Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention, 2020; Global Initiative for Asthma: Fontana, WI, USA, 2020. [Google Scholar]
- Reddel, H.K.; Taylor, D.R.; Bateman, E.D.; Boulet, L.P.; Boushey, H.A.; Busse, W.W.; Casale, T.B.; Chanez, P.; Enright, P.L.; Gibson, P.G.; et al. An official American Thoracic Society/European Respiratory Society statement: Asthma control and exacerbations—Standardizing endpoints for clinical asthma trials and clinical practice. Am. J. Respir. Crit. Care Med. 2009, 180, 59–99. [Google Scholar] [CrossRef] [Green Version]
- Dweik, R.A.; Boggs, P.B.; Erzurum, S.C.; Irvin, C.G.; Leigh, M.W.; Lundberg, J.O.; Olin, A.C.; Plummer, A.L.; Taylor, D.R.; on behalf of the American Thoracic Society Committee on Interpretation of Exhaled Nitric Oxide Levels (FeNO) for Clinical Applications. An official ATS clinical practice guideline: Interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am. J. Respir. Crit. Care Med. 2011, 184, 602–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jongste, J.C. Yes to NO: The first studies on exhaled nitric oxide-driven asthma treatment. Eur. Respir. J. 2005, 26, 379–381. [Google Scholar] [CrossRef]
- Smith, A.D.; Cowan, J.O.; Brassett, K.P.; Herbison, G.P.; Taylor, D.R. Use of Exhaled Nitric Oxide Measurements to Guide Treatment in Chronic Asthma. N. Engl. J. Med. 2005, 352, 2163–2173. [Google Scholar] [CrossRef] [Green Version]
- Pellegrino, R.; Viegi, G.; Brusasco, V.; Crapo, R.O.; Burgos, F.; Casaburi, R.; Coates, A.; van der Grinten, C.P.M.; Gustafsson, P.; Hankinson, J.; et al. Interpretative strategies for lung function tests. Eur. Respir. J. 2005, 26, 948–968. [Google Scholar] [CrossRef] [PubMed]
- Žuvela, P.; Lovric, M.; Yousefian-Jazi, A.; Liu, J.J. Ensemble Learning Approaches to Data Imbalance and Competing Objectives in Design of an Industrial Machine Vision System. Ind. Eng. Chem. Res. 2020, 59, 4636–4645. [Google Scholar] [CrossRef]
- Friedman, J.H. On bias, variance, 0/1-loss, and the curse-of-dimensionality. Data Min. Knowl. Discov. 1997, 1, 55–77. [Google Scholar] [CrossRef]
- Sheehan, W.J.; Phipatanakul, W. Indoor allergen exposure and asthma outcomes. Curr. Opin. Pediatr. 2016, 28, 772–777. [Google Scholar] [CrossRef] [Green Version]
- Lombardi, C.; Savi, E.; Ridolo, E.; Passalacqua, G.; Canonica, G.W. Is allergic sensitization relevant in severe asthma? Which allergens may be culprit? World Allergy Organ. J. 2017, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chicco, D. Ten quick tips for machine learning in computational biology. BioData Min. 2017, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar] [CrossRef]
- Zhang, Y.P.; Zhang, L.N.; Wang, Y.C. Cluster-based majority under-sampling approaches for class imbalance learning. In Proceedings of the 2010 2nd IEEE International Conference on Information and Financial Engineering (ICIFE 2010), Chongqing, China, 17–19 September 2010; pp. 400–404. [Google Scholar]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.I. Explainable AI for trees: From local explanations to global understanding. Nat. Mach. Intell. 2019, 2. [Google Scholar] [CrossRef] [PubMed]
- Freund, Y.; Schapire, R.E. Experiments with a New Boosting Algorithm. In Proceedings of the Thirteenth International Conference on Machine Learning, Bari, Italy, 3–6 July 1996; pp. 148–156. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Strobl, C.; Boulesteix, A.L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef] [Green Version]
- Havaš Auguštin, D.; Šarac, J.; Lovrić, M.; Živković, J.; Malev, O.; Fuchs, N.; Novokmet, N.; Turkalj, M.; Missoni, S.; Auguštin, D.H.; et al. Adherence to Mediterranean diet and maternal lifestyle during pregnancy: Island-mainland differentiation in the CRIBS birth cohort. Nutrients 2020, 12, 2179. [Google Scholar] [CrossRef]
- Lučić, B.; Batista, J.; Bojović, V.; Lovrić, M.; Sović Kržić, A.; Bešlo, D.; Nadramija, D.; Vikić-Topić, D. Estimation of Random Accuracy and its Use in Validation of Predictive Quality of Classification Models within Predictive Challenges. Croat. Chem. Acta 2019, 92. [Google Scholar] [CrossRef] [Green Version]
- Chicco, D.; Rovelli, C. Computational prediction of diagnosis and feature selection on mesothelioma patient health records. PLoS ONE 2019, 14, e0208737. [Google Scholar] [CrossRef]
- Weiss, G. Mining with rarity: A unifying framework. SIGKDD Explor. 2004, 6, 7–19. [Google Scholar] [CrossRef]
- Holzinger, A.; Biemann, C.; Pattichis, C.S.; Kell, D.B. What do we need to build explainable AI systems for the medical domain? arXiv 2017, arXiv:1712.09923. [Google Scholar]
- Holzinger, A. Interactive machine learning for health informatics: When do we need the human-in-the-loop? Brain Inform. 2016, 3, 119–131. [Google Scholar] [CrossRef] [Green Version]
- Loyola-Gonzalez, O. Black-box vs. White-Box: Understanding their advantages and weaknesses from a practical point of view. IEEE Access 2019, 7, 154096–154113. [Google Scholar] [CrossRef]
- Lovrić, M.; Pavlović, K.; Žuvela, P.; Spataru, A.; Lučić, B.; Kern, R.; Wong, M.W. Machine learning in prediction of intrinsic aqueous solubility of drug-like compounds: Generalization, complexity, or predictive ability? J. Chemom. 2021, e3349. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2000; Volume 7, ISBN 978-1-4614-7137-0. [Google Scholar]
- Harkins, M.S.; Fiato, K.L.; Iwamoto, G.K. Exhaled nitric oxide predicts asthma exacerbation. J. Asthma 2004, 41, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Camargo, C.A.; Faridi, M.K.; Yun, B.J.; Hasegawa, K. Machine learning approaches for predicting disposition of asthma and COPD exacerbations in the ED. Am. J. Emerg. Med. 2018, 36, 1650–1654. [Google Scholar] [CrossRef]
- Wang, R.; Simpson, A.; Custovic, A.; Foden, P.; Belgrave, D.; Murray, C.S. Individual risk assessment tool for school-age asthma prediction in UK birth cohort. Clin. Exp. Allergy 2019, 49, 292–298. [Google Scholar] [CrossRef]
- Lovrić, M.; Meister, R.; Steck, T.; Fadljević, L.; Gerdenitsch, J.; Schuster, S.; Schiefermüller, L.; Lindstaedt, S.; Kern, R. Parasitic resistance as a predictor of faulty anodes in electro galvanizing: A comparison of machine learning, physical and hybrid models. Adv. Model. Simul. Eng. Sci. 2020. [Google Scholar] [CrossRef]
- Celebi, R.; Uyar, H.; Yasar, E.; Gumus, O.; Dikenelli, O.; Dumontier, M. Evaluation of knowledge graph embedding approaches for drug-drug interaction prediction in realistic settings. BMC Bioinform. 2019, 20, 726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groningen, E.Z.; Kerstjens, H.A.M.; Brand, P.L.P.; Koeter, G.H.; Postma, D.S.; De Jong, P.M. Influence of treatment on peak expiratory flow and its relation to airway hyperresponsiveness and symptoms. Thorax 1994, 49, 1109–1115. [Google Scholar] [CrossRef] [Green Version]
- Brand, P.L.P.; Duiverman, E.J.; Waalkens, H.J.; Van Essen-Zandvliet, E.E.M.; Kerrebijn, K.F. Peak flow variation in childhood asthma: Correlation with symptoms, airways obstruction, and hyperresponsiveness during long term treatment with inhaled corticosteroids. Thorax 1999, 54, 103–107. [Google Scholar] [CrossRef] [Green Version]
- Tonascia, J.; Adkinson, N.F.; Bender, B.; Cherniack, R.; Donithan, M.; Kelly, H.W.; Reisman, J.; Shapiro, G.G.; Sternberg, A.L.; Strunk, R.; et al. Long-Term Effects of Budesonide or Nedocromil in Children with Asthma. N. Engl. J. Med. 2000, 343, 1054–1063. [Google Scholar] [CrossRef]
- Smith, A.D.; Cowan, J.O.; Brassett, K.P.; Filsell, S.; McLachlan, C.; Monti-Sheehan, G.; Herbison, C.P.; Taylor, D.R. Exhaled nitric oxide: A predictor of steroid response. Am. J. Respir. Crit. Care Med. 2005, 172, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Bagnasco, D.; Ferrando, M.; Varricchi, G.; Passalacqua, G.; Canonica, G.W. A critical evaluation of Anti-IL-13 and Anti-IL-4 strategies in severe asthma. Int. Arch. Allergy Immunol. 2016, 170, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Price, D.; Ryan, D.; Burden, A.; Von Ziegenweidt, J.; Gould, S.; Freeman, D.; Gruffydd-Jones, K.; Copland, A.; Godley, C.; Chisholm, A.; et al. Using fractional exhaled nitric oxide (FeNO) to diagnose steroid-responsive disease and guide asthma management in routine care. Clin. Transl. Allergy 2013, 3, 37. [Google Scholar] [CrossRef] [Green Version]
- Carrington, A.M.; Fieguth, P.W.; Qazi, H.; Holzinger, A.; Chen, H.H.; Mayr, F.; Manuel, D.G. A new concordant partial AUC and partial c statistic for imbalanced data in the evaluation of machine learning algorithms. BMC Med. Inform. Decis. Mak. 2020, 20, 4. [Google Scholar] [CrossRef]
- Belgrave, D.; Henderson, J.; Simpson, A.; Buchan, I.; Bishop, C.; Custovic, A. Disaggregating asthma: Big investigation versus big data. J. Allergy Clin. Immunol. 2017, 139, 400–407. [Google Scholar] [CrossRef] [Green Version]
- Bhakta, N.R.; Woodruff, P.G. Human asthma phenotypes: From the clinic, to cytokines, and back again. Immunol. Rev. 2011, 242, 220–232. [Google Scholar] [CrossRef]
- Froidure, A.; Mouthuy, J.; Durham, S.R.; Chanez, P.; Sibille, Y.; Pilette, C. Asthma phenotypes and IgE responses. Eur. Respir. J. 2016, 47, 304–319. [Google Scholar] [CrossRef] [Green Version]
- Hirano, T.; Matsunaga, K. Late-onset asthma: Current perspectives. J. Asthma Allergy 2018, 11, 19–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trivedi, M.; Denton, E. Asthma in children and adults—What are the differences and what can they tell us about asthma? Front. Pediatr. 2019, 7, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Freishtat, R.J.; Gordish-Dressman, H.; Teach, S.J.; Resca, L.; Hoffman, E.P.; Wang, Z. Natural progression of childhood asthma symptoms and strong influence of sex and puberty. Ann. Am. Thorac. Soc. 2014, 11, 898–907. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.; Bode, E.; Boer, L.; Hansel, T.T.; Blaser, K.; Johann-Christian Virchow, J. Allergic and Nonallergic Asthmatics Have Distinct Patterns of T-Cell Activation and Cytokine Production in Peripheral Blood and Bronchoalveolar Lavage. Am. Rev. Respir. Dis. 1992, 146, 109–115. [Google Scholar] [CrossRef] [PubMed]



| Variable Group | Description |
|---|---|
| demographics | gender, age |
| subjective clinical data | at baseline (t0)-personal and family medical history-atopy status, allergic rhinitis (AR), atopic dermatitis (AD), food allergy and other comorbidities |
| objective clinical data | at baseline (t0) and after 6 months (t0 + 6)-symptom control, frequency and severity of exacerbations in the period since the last visit, lung function (FVC, FEV1, MEF50), airway inflammation (FENO) measurement and medication use; at baseline (t0)- skin prick (SPT) and total and specific IgE to inhaled allergens, blood eosinophils and neutrophils, anthropometric measures (height, weight, body mass index) and for certain patients with suggestive history for comorbidities -ENT examination, pH probing with impedance for diagnostics of laryngopharyngeal reflux and gastroesophageal reflux disease, polysomnography for diagnostics of obstructive sleep apnea syndrome, SPT and specific IgE to food and insect venom allergens for diagnostics of food/insect venom allergy |
| genetic data | genotypes for rs37973 (GLCCI1), rs9910408 (TBX21), rs242941 (CRHR1), rs1876828 (CRHR1), rs1042713 (ADRB2) and rs17576 (MMP9) (see Table S1, Figure S1 and Table S2a,b in the Supplement) |
| Class | FEV1 | MEF50 | FENO | Asthma Control |
|---|---|---|---|---|
| Responders | Increase ≥ 10% predicted | Increase ≥ 15% predicted | Decrease < 20% for values > 35 (50) ppb or < 10 ppb for values < 35 (50) ppb | Improvement in asthma control |
| Non-Responders | Change < 10% predicted | Change < 15% predicted | Decrease ≤ 20% FENO ≤ 20% for values over 35 (50) ppb or ± 10 ppb for values < 35 (50) ppb or increase >20% for values > 35 (50) ppb or > 10 ppb for values < 35 (50) ppb | No changes in partial asthma control or deterioration in asthma control |
| Treatment Outcome (t0 + 6) | Responders (1) | Non-Responders (0) |
|---|---|---|
| LOAC (t0 + 6) | 230 | 135 |
| FENO (t0 + 6) | 248 | 104 |
| FEV1 (t0 + 6) | 129 | 236 |
| MEF50 (t0 + 6) | 126 | 239 |
| 1. Classification Algorithm | 2. Sampling Methods | 3. Targets After Six Months of Treatment |
|---|---|---|
| (a) AdaBoost | (a) No sampling (base) | (a) MEF50 (t0 + 6) |
| (b) Random Forest | (b) Under sampling (cluster centroids) | (b) FEV1 (t0 + 6) |
| (c) Oversampling | (c) FENO (t0 + 6) | |
| (d) LOAC (t0 + 6) |
| FEV1 | FENO | MEF50 | LOAC | |
|---|---|---|---|---|
| Accuracy | 0.6503 | 0.7005 | 0.6753 | 0.9698 |
| Specificity | 0.8986 | 0.8531 | 0.8817 | 0.9661 |
| Sensitivity | 0.7854 | 0.9560 | 0.7855 | 0.9781 |
| MCC | 0.2190 | 0.2146 | 0.2608 | 0.9366 |
| Variable | LOAC | FENO | FEV1 | MEF50 |
|---|---|---|---|---|
| Seasonal allergens (SPT) | 1.1% | |||
| Asthma severity (t0) | 47.0% | |||
| hsCRP | 1.2% | |||
| IgE total | 1.5% | 3.2% | ||
| FENO (t0) | 12.8% | |||
| FEV1 (t0) | 14.8% | 1.8% | ||
| MEF50 (t0) | 8.2% | 30.3% |
| Response | Age Group | MCC (All Models) | No. Patients |
|---|---|---|---|
| LOAC | 2–5 y/o | b 1 | 53 |
| 6–11 y/o | 0.96 | 178 | |
| 12–17 y/o | 0.89 | 124 | |
| >18 y/o | w 0.61 | 10 | |
| FENO | 2–5 y/o | w 0 | 53 |
| 6–11 y/o | b 0.14 | 178 | |
| 12–17 y/o | 0.1 | 124 | |
| >18 y/o | w 0 | 10 | |
| FEV1 | 2–5 y/o | 0.12 | 53 |
| 6–11 y/o | b 0.27 | 178 | |
| 12–17 y/o | 0.08 | 124 | |
| >18 y/o | w 0 | 10 | |
| MEF50 | 2–5 y/o | b 0.29 | 53 |
| 6–11 y/o | 0.18 | 178 | |
| 12–17 y/o | 0.25 | 124 | |
| >18 y/o | w 0 | 10 |
| Response | Sampling | Logistic Regression | AdaBoost | Random Forest |
|---|---|---|---|---|
| FENO | Base | 0.07 | 0.21 | 0.07 |
| CC | −0.02 | 0.07 | 0.10 | |
| OS | 0.05 | 0.17 | 0.13 | |
| FEV1 | Base | 0.00 | 0.22 | 0.14 |
| CC | 0.03 | 0.14 | 0.15 | |
| OS | 0.03 | 0.18 | 0.19 | |
| LOAC | Base | 0.19 | 0.94 | 0.90 |
| CC | 0.04 | 0.89 | 0.90 | |
| OS | 0.17 | 0.93 | 0.90 | |
| MEF50 | Base | −0.01 | 0.23 | 0.19 |
| CC | −0.02 | 0.14 | 0.12 | |
| OS | 0.03 | 0.24 | 0.26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lovrić, M.; Banić, I.; Lacić, E.; Pavlović, K.; Kern, R.; Turkalj, M. Predicting Treatment Outcomes Using Explainable Machine Learning in Children with Asthma. Children 2021, 8, 376. https://0-doi-org.brum.beds.ac.uk/10.3390/children8050376
Lovrić M, Banić I, Lacić E, Pavlović K, Kern R, Turkalj M. Predicting Treatment Outcomes Using Explainable Machine Learning in Children with Asthma. Children. 2021; 8(5):376. https://0-doi-org.brum.beds.ac.uk/10.3390/children8050376
Chicago/Turabian StyleLovrić, Mario, Ivana Banić, Emanuel Lacić, Kristina Pavlović, Roman Kern, and Mirjana Turkalj. 2021. "Predicting Treatment Outcomes Using Explainable Machine Learning in Children with Asthma" Children 8, no. 5: 376. https://0-doi-org.brum.beds.ac.uk/10.3390/children8050376