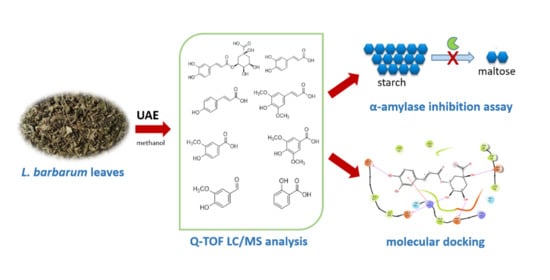
Phenolic Acids from Lycium barbarum Leaves: In Vitro and In Silico Studies of the Inhibitory Activity against Porcine Pancreatic α-Amylase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Sample
2.2. Reagents
2.3. Ultrasound-Assisted Extraction (UAE) of L. barbarum Leaves
2.4. Determination of Total Phenol Content (TPC)
2.5. In Vitro Antioxidant Activities
2.5.1. Free Radical-Scavenging Activity Using DPPH Assay
2.5.2. Free Radical-Scavenging Activity Using ABTS Assay
2.5.3. Ferric Reducing Antioxidant Power (FRAP) Assay
2.6. α-Amylase Inhibition Activity Assay
2.7. Analysis of Phenolic Acids by Q-TOF-LC/MS
2.8. Molecular Docking
2.9. Statistical Analysis
3. Results and Discussion
3.1. Characterization of L. barbarum Leaf Extract and α-Amylase Inhibition of Constituent Phenolic Acids
3.2. Docking Studies
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blasi, F.; Urbani, E.; Simonetti, M.S.; Chiesi, C.; Cossignani, L. Seasonal variations in antioxidant compounds of Olea europaea leaves collected from different Italian cultivars. J. Appl. Bot. Food Qual. 2016, 89, 202–207. [Google Scholar]
- Durazzo, A.; D’Addezio, L.; Camilli, E.; Piccinelli, R.; Turrini, A.; Marletta, L.; Marconi, S.; Lucarini, M.; Lisciani, S.; Gabrielli, P.; et al. From Plant Compounds to Botanicals and Back: A Current Snapshot. Molecules 2018, 23, 1844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montesano, D.; Montesano, D.; Giua, L.; Urbani, E.; Simonetti, M.S.; Blasi, F. A Simple HPLC-ELSD Method for Sugar Analysis in Goji Berry. J. Chem. 2016, 2016, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Blasi, F.; Montesano, D.; Simonetti, M.S.; Cossignani, L. A Simple and Rapid Extraction Method to Evaluate the Fatty Acid Composition and Nutritional Value of Goji Berry Lipid. Food Anal. Methods 2016, 10, 970–979. [Google Scholar] [CrossRef]
- Cossignani, L.; Cossignani, L.; Blasi, F.; Perini, M.; Barbero, A.; Pianezze, S.; Montesano, D. Characterisation and geographical traceability of Italian goji berries. Food Chem. 2019, 275, 585–593. [Google Scholar] [CrossRef]
- Cossignani, L.; Blasi, F.; Simonetti, M.S.; Montesano, D. Fatty Acids and Phytosterols to Discriminate Geographic Origin of Lycium barbarum Berry. Food Anal. Methods 2017, 11, 1180–1188. [Google Scholar] [CrossRef]
- Pollini, L.; Rocchi, R.; Cossignani, L.; Mañes, J.; Compagnone, D.; Blasi, F. Phenol Profiling and Nutraceutical Potential of Lycium spp. Leaf Extracts Obtained with Ultrasound and Microwave Assisted Techniques. Antioxidants 2019, 8, 260. [Google Scholar] [CrossRef] [Green Version]
- Mocan, A.; Zengin, G.; Simirgiotis, M.; Schafberg, M.; Mollica, A.; Vodnar, D.C.; Crişan, G.; Rohn, S. Functional constituents of wild and cultivated Goji (L. barbarum L.) leaves: Phytochemical characterization, biological profile, and computational studies. J. Enzym. Inhib. Med. Chem. 2017, 32, 153–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.-N.; Li, S.; Zhang, Y.-J.; Xu, X.-R.; Chen, Y.-M.; Li, H.-B. Resources and Biological Activities of Natural Polyphenols. Nutrients 2014, 6, 6020–6047. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yang, W.; Sun, W.; Chen, S.; Liu, D.; Kong, X.; Tian, J.; Ye, X. Inhibition of porcine pancreatic α-amylase activity by chlorogenic acid. J. Funct. Foods 2020, 64, 103587. [Google Scholar] [CrossRef]
- Gachons, C.P.D.; Breslin, P.A.S. Salivary Amylase: Digestion and Metabolic Syndrome. Curr. Diabetes Rep. 2016, 16, 102. [Google Scholar] [CrossRef]
- Dall’Acqua, S.; Locatelli, M.; Stefanucci, A.; Macedonio, G.; Novellino, E.; Mirzaie, S.; Dvorácskó, S.; Carradori, S.; Brunetti, L.; Orlando, G.; et al. Chemical characterization, antioxidant properties, anti-inflammatory activity and enzyme inhibition of Ipomoea batatas L. leaf extracts. Int. J. Food Prop. 2017, 20, 1907–1919. [Google Scholar] [CrossRef] [Green Version]
- Mahomoodally, M.F.; Mollica, A.; Stefanucci, A.; Aumeeruddy, M.Z.; Poorneeka, R.; Dall’Acqua, S. Volatile components, pharmacological profile, and computational studies of essential oil from Aegle marmelos (Bael) leaves: A functional approach. Ind. Crop Prod. 2018, 126, 13–21. [Google Scholar] [CrossRef]
- Tomczyk, M.; Miłek, M.; Sidor, E.; Kapusta, I.; Litwińczuk, W.; Puchalski, C.; Dżugan, M. The Effect of Adding the Leaves and Fruits of Morus alba to Rape Honey on Its Antioxidant Properties, Polyphenolic Profile, and Amylase Activity. Molecules 2019, 25, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelli, I.; Benariba, N.; Adjdir, S.; Fekhikher, Z.; Daoud, I.; Terki, M.; Benramdane, H.; Ghalem, S. In silico evaluation of phenolic compounds as inhibitors of A-amylase and A-glucosidase. J. Biomol. Struct. Dyn. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Demir, Y.; Durmaz, L.; Taslimi, P.; Gülçin, I. Antidiabetic properties of dietary phenolic compounds: Inhibition effects on α-amylase, aldose reductase, and α-glycosidase. Biotechnol. Appl. Biochem. 2019, 66, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Pagano, C.; Perioli, L.; Blasi, F.; Bastianini, M.; Chiesi, C.; Cossignani, L. Optimisation of phenol extraction from wine using layered double hydroxides and technological evaluation of the bioactive-rich powder. Int. J. Food Sci. Technol. 2017, 52, 2582–2588. [Google Scholar] [CrossRef]
- Pollini, L.; Tringaniello, C.; Ianni, F.; Blasi, F.; Mañes, J.; Montesano, D. Impact of Ultrasound Extraction Parameters on the Antioxidant Properties of Moringa oleifera Leaves. Antioxidants 2020, 9, 277. [Google Scholar] [CrossRef]
- Urbani, E.; Blasi, F.; Simonetti, M.S.; Chiesi, C.; Cossignani, L. Investigation on secondary metabolite content and antioxidant activity of commercial saffron powder. Eur. Food Res. Technol. 2016, 242, 987–993. [Google Scholar] [CrossRef]
- Rocchetti, G.; Blasi, F.; Montesano, D.; Ghisoni, S.; Marcotullio, M.C.; Sabatini, S.; Cossignani, L.; Lucini, L. Impact of conventional/non-conventional extraction methods on the untargeted phenolic profile of Moringa oleifera leaves. Food Res. Int. 2019, 115, 319–327. [Google Scholar] [CrossRef]
- Wu, L.; Liu, Y.; Qin, Y.; Wang, L.; Wu, Z. HPLC-ESI-qTOF-MS/MS Characterization, Antioxidant Activities and Inhibitory Ability of Digestive Enzymes with Molecular Docking Analysis of Various Parts of Raspberry (Rubus ideaus L.). Antioxidants 2019, 8, 274. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Available online: http://pubchem.ncbi.nlm.nih.gov/ (accessed on 2 August 2020).
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein−Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2017, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Moscatello, S.; Frioni, T.; Blasi, F.; Proietti, S.; Pollini, L.; Verducci, G.; Rosati, A.; Walker, R.P.; Battistelli, A.; Cossignani, L.; et al. Changes in Absolute Contents of Compounds Affecting the Taste and Nutritional Properties of the Flesh of Three Plum Species Throughout Development. Foods 2019, 8, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mocan, A.; Vlase, L.; Vodnar, D.C.; Bischin, C.; Hanganu, D.; Gheldiu, A.-M.; Oprean, R.; Silaghi-Dumitrescu, R.; Crişan, G. Polyphenolic Content, Antioxidant and Antimicrobial Activities of Lycium barbarum L. and Lycium chinense Mill. Leaves. Molecules 2014, 19, 10056–10073. [Google Scholar] [CrossRef]
- Potterat, O. Goji (Lycium barbarum and L. chinense): Phytochemistry, Pharmacology and Safety in the Perspective of Traditional Uses and Recent Popularity. Planta Medica 2009, 76, 7–19. [Google Scholar] [CrossRef] [Green Version]
- Qian, D.; Zhao, Y.; Yang, G.; Huang, L. Systematic Review of Chemical Constituents in the Genus Lycium (Solanaceae). Molecules 2017, 22, 911. [Google Scholar] [CrossRef] [Green Version]
- Inbaraj, B.S.; Lu, H.; Kao, T.; Chen, B. Simultaneous determination of phenolic acids and flavonoids in Lycium barbarum Linnaeus by HPLC–DAD–ESI-MS. J. Pharm. Biomed. Anal. 2010, 51, 549–556. [Google Scholar] [CrossRef]
- Mocan, A.; Vlase, L.; Raita, O.; Hanganu, D.; Păltinean, R.; Dezsi, Ș.; Gheldiu, A.M.; Oprean, R.; Crisan, G. Comparative studies on antioxidant activity and polyphenolic content of Lycium barbarum L. and Lycium chinense Mill. leaves. Pak. J. Pharm. Sci. 2015, 28, 1511–1515. [Google Scholar] [PubMed]
- Min, S.W.; Han, J.S. Polyopes lancifolia Extract, a Potent α-Glucosidase Inhibitor, Alleviates Postprandial Hyperglycemia in Diabetic Mice. Prev. Nutr. Food Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funke, I.; Melzig, M.F. Effect of different phenolic compounds on α-amylase activity: Screening by microplate-reader based kinetic assay. Pharmazie 2005, 60, 796–797. [Google Scholar] [PubMed]
- Thummajitsakul, S.; Samaikam, S.; Tacha, S.; Silprasit, K. Study on FTIR spectroscopy, total phenolic content, antioxidant activity and anti-amylase activity of extracts and different tea forms of Garcinia schomburgkiana leaves. LWT 2020, 134, 110005. [Google Scholar] [CrossRef]
- Mocan, A.; Moldovan, C.; Zengin, G.; Bender, O.; Locatelli, M.; Simirgiotis, M.J.; Atalay, A.; Vodnar, D.C.; Rohn, S.; Crișan, G. UHPLC-QTOF-MS analysis of bioactive constituents from two Romanian Goji (Lycium barbarum L.) berries cultivars and their antioxidant, enzyme inhibitory, and real-time cytotoxicological evaluation. Food Chem. Toxicol. 2018, 115, 414–424. [Google Scholar] [CrossRef]
- Sachan, A.K.; Rao, C.V.; Sachan, N.K. In vitro studies on the inhibition of α-amylase and α-glucosidase by hydro-ethanolic extract of Pluchea lanceolata, Alhagi pseudalhagi, Caesalpinia bonduc. Pharmacogn. Res. 2019, 11, 310. [Google Scholar] [CrossRef]
- Jongkees, S.A.; Caner, S.; Tysoe, C.; Brayer, G.D.; Withers, S.G.; Suga, H. Rapid Discovery of Potent and Selective Glycosidase-Inhibiting De Novo Peptides. Cell Chem. Biol. 2017, 24, 381–390. [Google Scholar] [CrossRef]







| n. | Compounds | Concentration (µg/g) | Rt (min) | Mass Calculated (m/z, M-H−) | MS Fragments (m/z, M-H−) | Error (ppm) | Score |
|---|---|---|---|---|---|---|---|
| 1 | Syringic acid | 0.76 ± 0.000 | 4.39 | 197.0458 | 123.0083/147.8891/95.0138 | −1.22 | 98.26 |
| 2 | Chlorogenic acid | 358.34 ± 0.004 | 4.69 | 354.0955 | 191.0543/127.0397/85.0301 | −1.22 | 98.72 |
| 3 | Salicylic acid | 239.02 ± 0.005 | 5.13 | 137.0247 | 93.0351/65.0405 | −1.23 | 99.41 |
| 4 | Caffeic acid | 0.07 ± 0.000 | 5.67 | 180.0432 | 135.0447/107.0506/89.0404 | −1.10 | 97.22 |
| 5 | Vanillic acid | 9.46 ± 0.002 | 6.39 | 167.0350 | 108.0218/123.0458/91.0196 | −1.34 | 97.11 |
| 6 | p-Coumaric acid | 0.84 ± 0.000 | 6.58 | 223.0619 | 164,0483/119.0497/93.0349 | −1.50 | 97.23 |
| 7 | Sinapic acid | 2.36 ± 0.000 | 6.69 | 283.0833 | 224.0696/149.0232/93.0350 | −1.71 | 99.34 |
| 8 | Vanillin | 8.62 ± 0.003 | 4.47 | 197.0457 | 146.7437/136.016/76.9614 | −1.67 | 96.44 |
| n. | Compounds | Regression Equation | R2 | IC50 mg/mL | |
|---|---|---|---|---|---|
| Slope | Intercept | ||||
| 1 | Syringic acid | 46.400 | −228.280 | 0.9925 | 6.0 |
| 2 | Chlorogenic acid | 178.560 | −37.986 | 0.9987 | 0.5 |
| 3 | Salicylic acid | 384.000 | −633.930 | 0.9930 | 1.8 |
| 4 | Caffeic acid | 15.923 | −5.776 | 0.9995 | 3.5 |
| 5 | Vanillic acid | 12.512 | −27.403 | 0.9996 | 6.2 |
| 6 | p-Coumaric acid | 10.627 | −9.487 | 0.9989 | 5.6 |
| 7 | Sinapic acid | 8.863 | −23.288 | 0.9999 | 8.3 |
| 8 | Vanillin | 4.808 | −1.849 | 0.9990 | 10.8 |
| Acarbose | 0.553 | -18.057 | 0.9980 | 0.1 | |
| L. barbarum leaf extract | 2.185 | −5.520 | 0.9977 | 25.4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pollini, L.; Riccio, A.; Juan, C.; Tringaniello, C.; Ianni, F.; Blasi, F.; Mañes, J.; Macchiarulo, A.; Cossignani, L. Phenolic Acids from Lycium barbarum Leaves: In Vitro and In Silico Studies of the Inhibitory Activity against Porcine Pancreatic α-Amylase. Processes 2020, 8, 1388. https://0-doi-org.brum.beds.ac.uk/10.3390/pr8111388
Pollini L, Riccio A, Juan C, Tringaniello C, Ianni F, Blasi F, Mañes J, Macchiarulo A, Cossignani L. Phenolic Acids from Lycium barbarum Leaves: In Vitro and In Silico Studies of the Inhibitory Activity against Porcine Pancreatic α-Amylase. Processes. 2020; 8(11):1388. https://0-doi-org.brum.beds.ac.uk/10.3390/pr8111388
Chicago/Turabian StylePollini, Luna, Alessandra Riccio, Cristina Juan, Carmela Tringaniello, Federica Ianni, Francesca Blasi, Jordi Mañes, Antonio Macchiarulo, and Lina Cossignani. 2020. "Phenolic Acids from Lycium barbarum Leaves: In Vitro and In Silico Studies of the Inhibitory Activity against Porcine Pancreatic α-Amylase" Processes 8, no. 11: 1388. https://0-doi-org.brum.beds.ac.uk/10.3390/pr8111388