Not Just Numbers: Mathematical Modelling and Its Contribution to Anaerobic Digestion Processes
Abstract
:1. Introduction
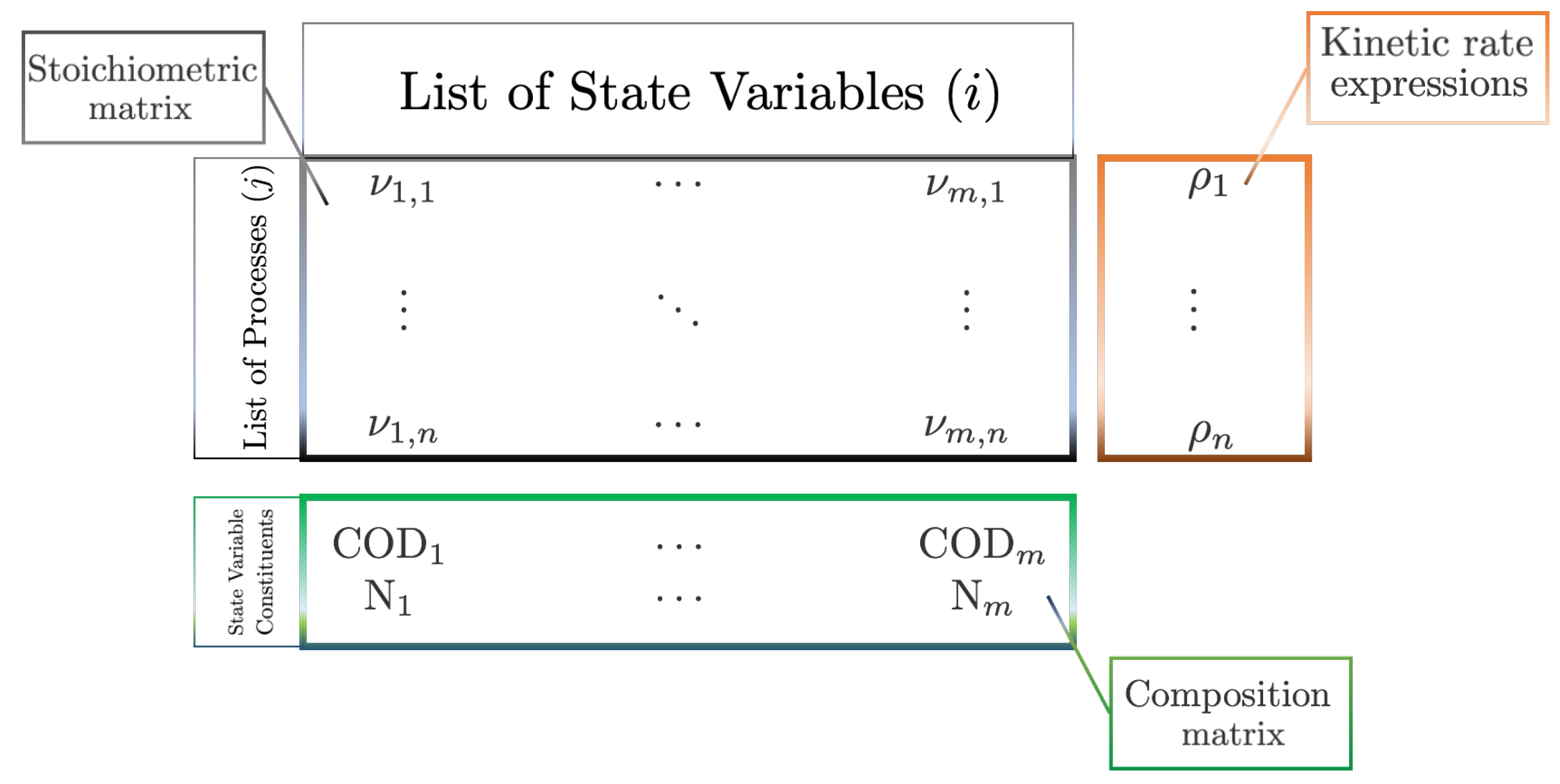
2. A First-Principles, Engineering Approach to Modelling Anaerobic Digestion
3. Reduced-Order Models Unlock the Power of Mathematics
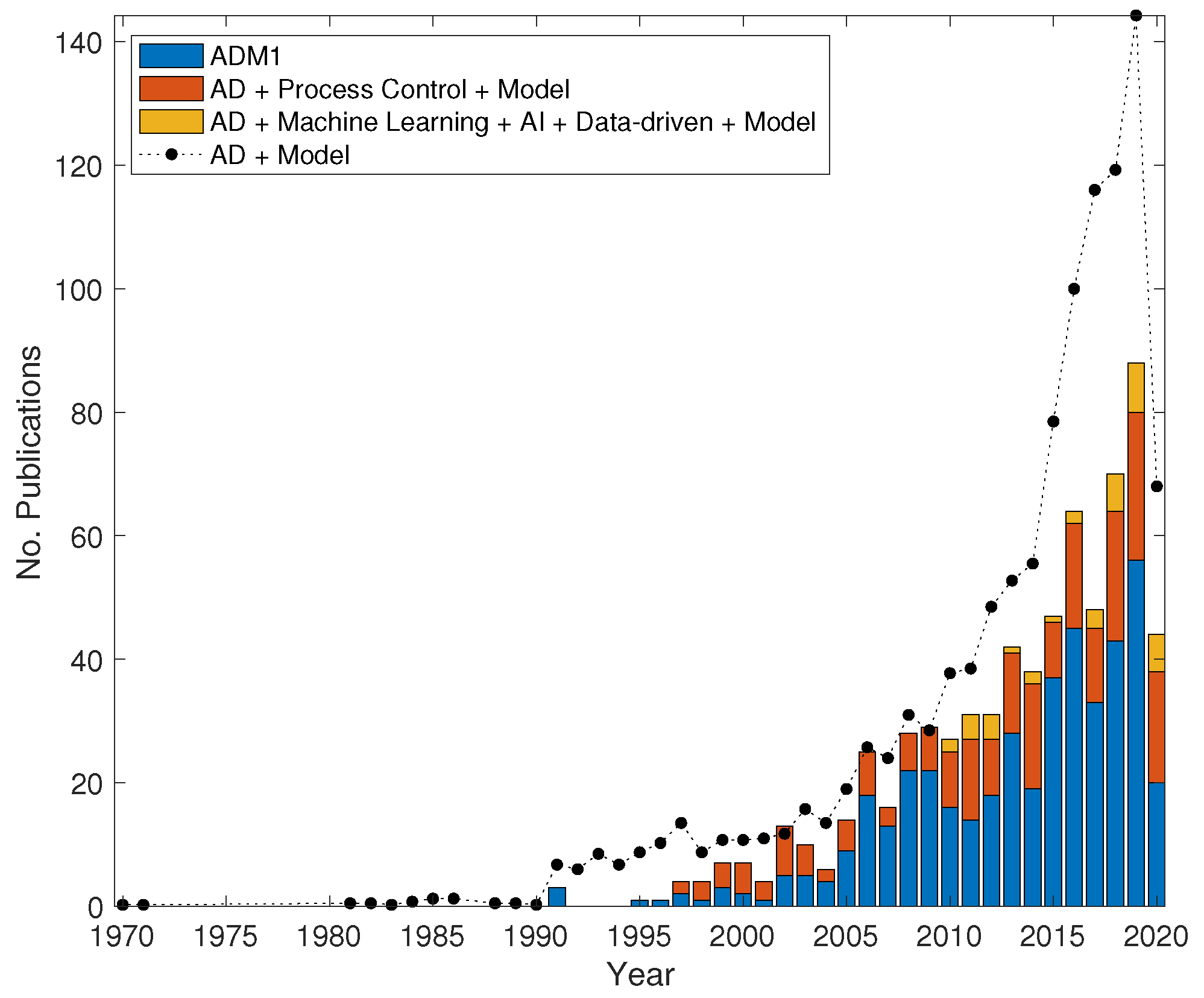
4. Empiricism, Data and the Digital Future
5. Multi-Disciplinarity and the Future of Anaerobic Digestion Modelling
5.1. Hybrid Models
Fundamental biology should not choose between small-scale mechanistic understanding and large-scale prediction. It should embrace the complementary strengths of mechanistic modelling and machine learning approaches …
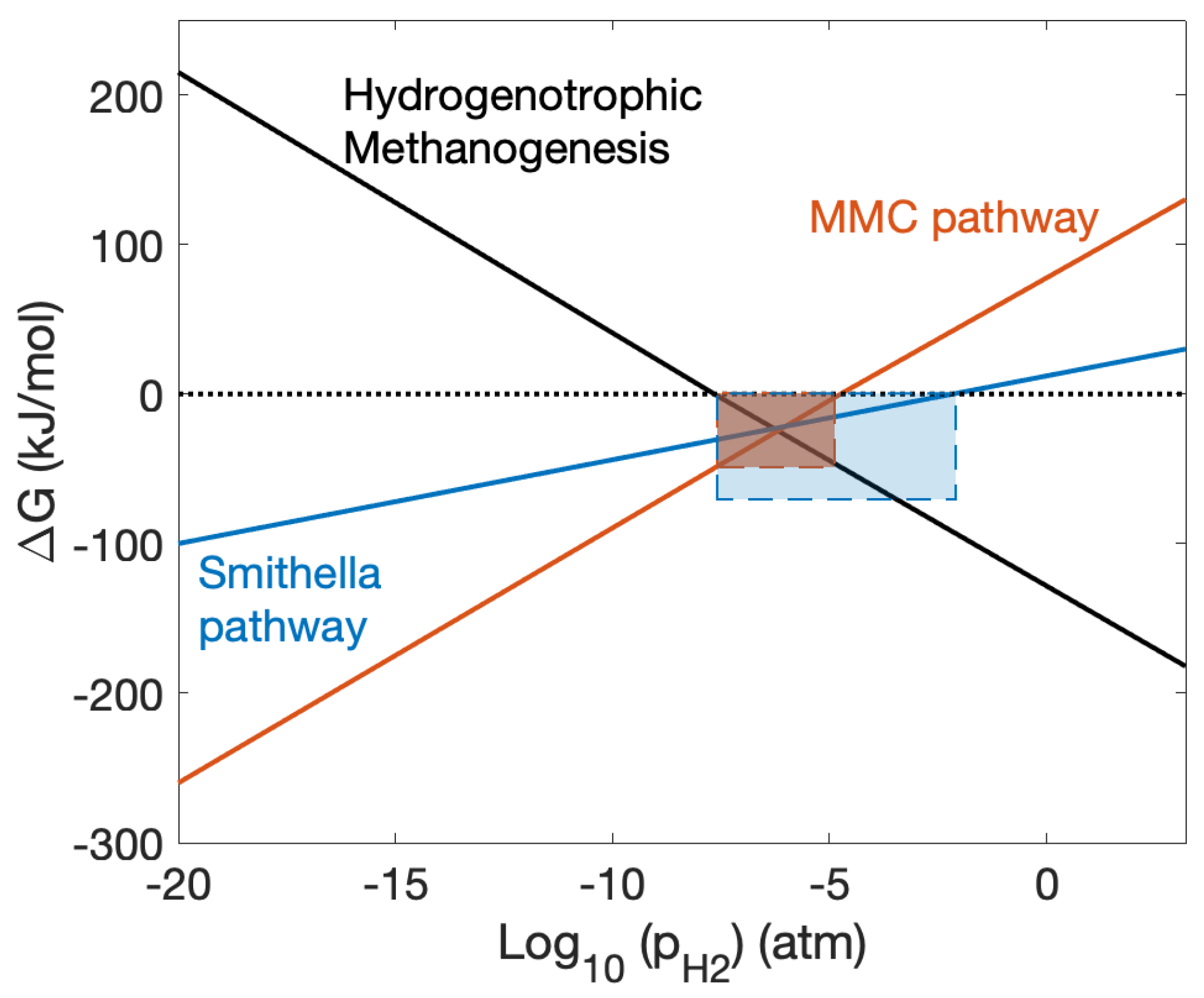
5.2. Thermodynamic Modelling
5.3. Computational Fluid Dynamics
5.4. Individual-Based Modelling
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Anaerobic Digestion |
| ADM1 | Anaerobic Digestion Model No. 1 |
| AI | Artificial Intelligence |
| ANN | Artificial Neural Network |
| ASMx | Activated Sludge Model No. (x) |
| BOD | Biochemical Oxygen Demand |
| CA | Cellular Automata |
| CFD | Computational Fluid Dynamics |
| COD | Chemical Oxygen Demand |
| DAE | Differential Algebraic Equations |
| EBS | Engineered Biological Systems |
| FBA | Flux Balance Analysis |
| IbM | Individual-based Model |
| ODE | Ordinary Differential Equation |
| OPLS | Ordinary Partial Least Squares |
| PCA | Principal Component Analysis |
| PDE | Partial Differential Equation |
| PLS | Partial Least Squares |
| VFA | Volatile Fatty Acids |
| VSS | Volatile Suspended Solids |
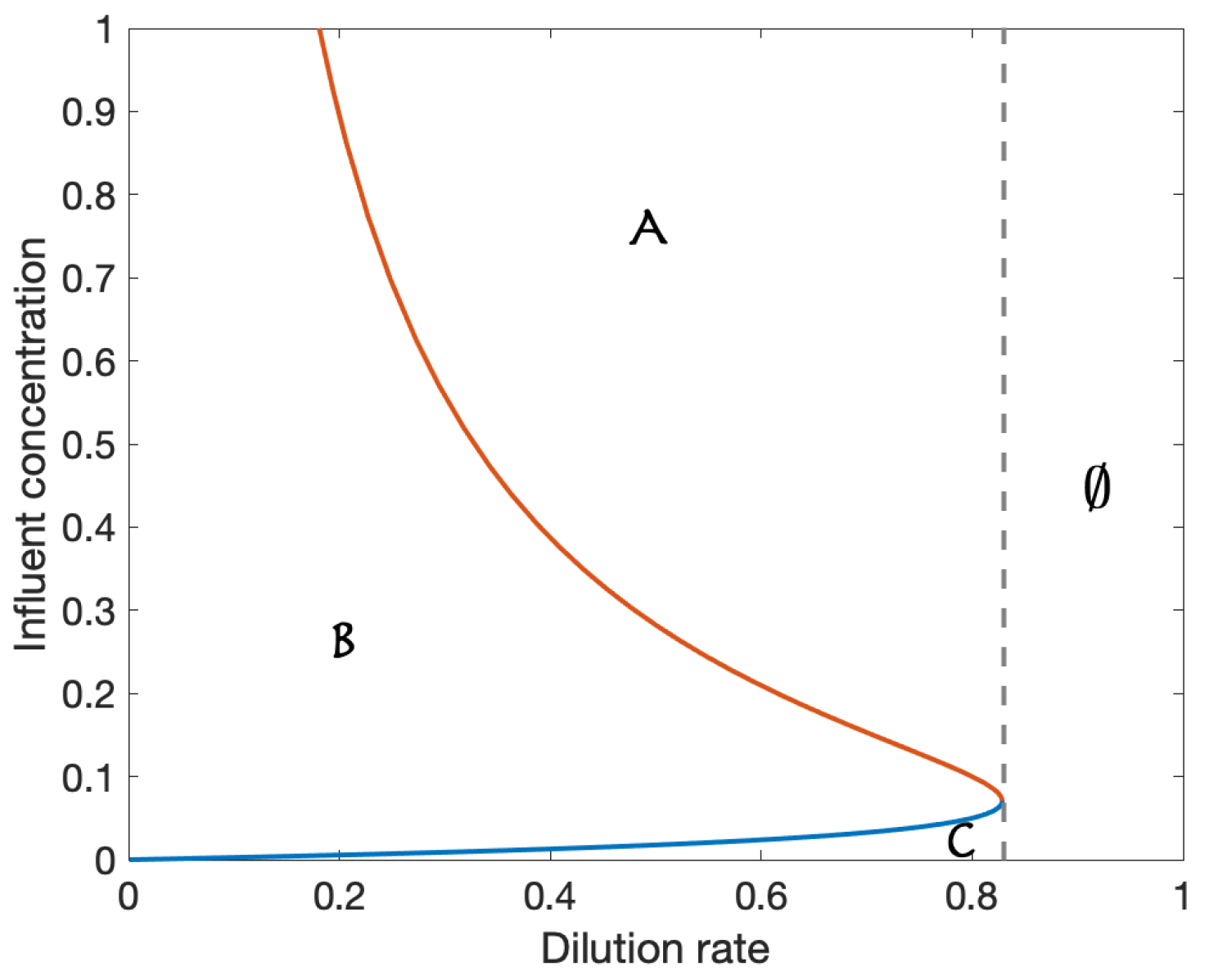
Appendix A. Bifurcation Analysis Example
| Parameter | Value |
|---|---|
| 2.0 | |
| Y | 0.5 |
| 0.05 | |
| 0.1 | |
| 0.5 | |
| D | 0.4 |

References
- Makinia, J. Mathematical Modelling and Computer Simulation of Activated Sludge Systems; IWA Publishing: London, UK, 2010. [Google Scholar]
- van Loosdrecht, M.C.M.; Lopez-Vazquez, C.M.; Meijer, S.C.F.; Hooijmans, C.M.; Brdjanovic, D. Twenty-five years of ASM1: Past, present and future of wastewater treatment modelling. J. Hydroinform. 2015, 17, 697–718. [Google Scholar] [CrossRef] [Green Version]
- Henze, M.; Grady, J.R.C.P.; Gujer, W.; Marais, G.; Matsuo, T. Activated Sludge Model No 1; IAWPRC Scientific and Technical Report No. 1; IAWQ: London, UK, 1987. [Google Scholar]
- Henze, M.; Gujer, W.; Mino, T.; Matsuo, T.; Wentzel, M.C.; Marais, G.v.R. Activated Sludge Model No. 2; IAWQ Scientific and Technical Report No. 3; IAWQ: London, UK, 1995. [Google Scholar]
- Henze, M.; Gujer, W.; Mino, T.; Matsuo, T.; Wentzel, M.C.; Marais, G.v.R.; van Loosdrecht, M.C.M. Activated Sludge Model No. 2d, ASM2d. Water Sci. Technol. 1999, 39, 165–182. [Google Scholar] [CrossRef]
- Batstone, D.J.; Keller, J.; Angelidaki, I.; Kalyuzhnyi, S.V.; Pavlostathis, S.G.; Rozzi, A.; Sanders, W.T.M.; Siegrist, H.; Vavilin, V.A. Anaerobic Digestion Model No. 1; Technical Report Report No. 13; IWA Publishing: London, UK, 2002. [Google Scholar]
- Jeppsson, U.; Rosen, C.; Alex, J.; Copp, J.B.; Gernaey, K.V.; Pons, M.N.; Vanrolleghem, P.A. Towards a benchmark simulation model for plant-wide control strategy performance evaluation of WWTPs. Water Sci. Technol. 2006, 53, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Jeppsson, U.; Pons, M.N.; Nopens, I.; Alex, J.; Copp, J.B.; Gernaey, K.V.; Rosen, C.; Steyer, J.P.; Vanrolleghem, P.A. Benchmark Simulation Model No. 2—General protocol and exploratory case studies. Water Sci. Technol. 2007, 56, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Rosen, C.; Vrecko, D.; Gernaey, K.; Pons, M.; Jeppsson, U. Implementing ADM1 for plant-wide benchmark simulations in Matlab/Simulink. Water Sci. Technol. 2006, 54, 11–19. [Google Scholar] [CrossRef]
- Arnell, M.; Rahmberg, M.; Oliveira, F.; Jeppsson, U. Multi-objective performance assessment of wastewater treatment plants combining plant-wide process models and life cycle assessment. J. Water Clim. Chang. 2017, 8, 715–729. [Google Scholar] [CrossRef]
- Fernández-Arévalo, T.; Lizarralde, I.; Fdz-Polanco, F.; Pérez-Elvira, S.I.; Garrido, J.M.; Puig, S.; Poch, M.; Grau, P.; Ayesa, E. Quantitative assessment of energy and resource recovery in wastewater treatment plants based on plant-wide simulations. Water Res. 2017, 118, 272–288. [Google Scholar] [CrossRef]
- Solon, K.; Mingsheng, J.; Volcke, E.I.P. Process schemes for future energy-positive water resource recovery facilities. Water Sci. Technol. 2019, 79, 1808–1820. [Google Scholar] [CrossRef]
- Maere, T.; Verrecht, B.; Moerenhout, S.; Judd, S.; Nopens, I. BSM-MBR: A benchmark simulation model to compare control and operational strategies for membrane bioreactors. Water Res. 2011, 45, 2181–2190. [Google Scholar] [CrossRef] [Green Version]
- Seco, A.; Ruano, M.V.; Ruiz-Martinez, A.; Robles, A.; Barat, R.; Serralta, J.; Ferrer, J. Plant-wide modelling in wastewater treatment: Showcasing experiences using the Biological Nutrient Removal Model. Water Sci. Technol. 2020, 81, 1700–1714. [Google Scholar] [CrossRef]
- Fedorovich, V.; Lens, P.; Kalyuzhnyi, S. Extension of Anaerobic Digestion Model No. 1 with processes of sulfate reduction. Appl. Biochem. Biotechnol. 2003, 109, 33–45. [Google Scholar] [CrossRef]
- Batstone, D.J.; Keller, J.; Steyer, J.P. A review of ADM1 extensions, applications, and analysis: 2002–2005. Water Sci. Technol. 2006, 54, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Flores-Alsina, X.; Solon, K.; Mbamba, C.K.; Tait, S.; Gernaey, K.V.; Jeppsson, U.; J.Batstone, D. Modelling phosphorus (P), sulfur (S) and iron (Fe) interactions for dynamic simulations of anaerobic digestion processes. Water Res. 2016, 95, 370–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frunzo, L.; Fermoso, F.G.; Luongo, V.; Mattei, M.R.; Esposito, G. ADM1-based mechanistic model for the role of trace elements in anaerobic digestion processes. J. Environ. Manag. 2019, 241, 587–602. [Google Scholar] [CrossRef]
- Siegrist, H.; Vogt, D.; Garcia-Heras, J.; Gujer, W. Mathematical model for meso and thermophilic anaerobic sewage sludge digestion. Environ. Sci. Technol. 2002, 36, 1113–1123. [Google Scholar] [CrossRef]
- Elmitwalli, T.A.; Sayed, S.; Groendijk, L.; van Lier, J.; Zeeman, G.; Lettinga, G. Decentralised treatment of concentrated sewage at low temperature in a two-step anaerobic system: Two upflow-hybrid septic tanks. Water Sci. Technol. 2003, 48, 219–226. [Google Scholar] [CrossRef]
- Copp, J.; Peerbolte, A.; Snowling, S.; Schraa, O.; Froelich, D.; Belia, E. Integrating anaerobic digestion into plant-wide wastewater treatment modelling—Experience with data from a large treatment plant. In Proceedings of the Anaerobic Digestion 2004, 10th World Congress on Anaerobic Digestion, Montréal, QC, Canada, 29 August–2 September 2004; pp. 1362–1365. [Google Scholar]
- Batstone, D.J.; Puyol, D.; Flores-Alsina, X.; Rodríguez, J. Mathematical modelling of anaerobic digestion processes: Applications and future needs. Rev. Environ. Sci. Biotechnol. 2015, 14, 595–613. [Google Scholar] [CrossRef]
- Arzate, J.A.; Kirstein, M.; Ertem, F.C.; Kielhorn, E.; Ramirez Malule, H.; Neubauer, P.; Cruz-Bournazou, M.N.; Junne, S. Anaerobic Digestion Model (AM2) for the Description of Biogas Processes at Dynamic Feedstock Loading Rates. Chem. Ing. Tech. 2017, 89, 686–695. [Google Scholar] [CrossRef]
- Hoffmann, D.S. The dawn of mathematical biology. arXiv 2015, arXiv:physics.hist-ph/1511.01455. [Google Scholar]
- Michaelis, L.; Menten, M.L. Die Kinetik der Invertinwirkung [The kinetics of invertase activity]. Biochem. Z. 1913, 49, 333–369. [Google Scholar]
- Israel, G. On the Contribution of Volterra and Lotka to the Development of Modern Biomathematics. Hist. Philos. Life Sci. 1988, 10, 37–49. [Google Scholar] [PubMed]
- Monod, J. Recherches sur la Croissance des Cellules Bactériennes [Research on the Growth of Bacterial Cells]. Ph.D. Thesis, Université de Paris, Paris, France, 1941. [Google Scholar]
- Wade, M.J.; Harmand, J.; Benyahia, B.; Bouchez, T.; Chaillou, S.; Cloez, B.; Godon, J.; Moussa Boudjemaa, B.; Rapaport, A.; Sari, T.; et al. Perspectives in mathematical modelling for microbial ecology. Ecol. Modell. 2016, 321, 64–74. [Google Scholar] [CrossRef]
- Andrews, J.F. A Mathematical Model for the Continuous Culture of Microorganisms Utilizing Inhibitory Substrate. Biotechnol. Bioeng. 1968, 10, 707–723. [Google Scholar] [CrossRef]
- Andrews, J.F. Dynamic model of the anaerobic digestion process. J. Sanit. Eng. Div. Am. Soc. Civ. Eng. 1969, 95, 95–116. [Google Scholar]
- Lawrence, A.W.; McCarty, P.L. Kinetics of methane fermentation in anaerobic treatment. J. Water Pollut. Control Fed. 2009, 41, R1–R17. [Google Scholar]
- Lawrence, A.W. Application of Process Kinetics to Design of Anaerobic Processes. In Anaerobic Biological Treatment Processes; American Chemical Society: Washington, DC, USA, 1971; Volume 105, pp. 163–190. [Google Scholar] [CrossRef]
- Andrews, J.F.; Graef, S.P. Dynamic modelling and Simulation of the Anaerobic Digestion Process. In Anaerobic Biological Treatment Processes; American Chemical Society: Washington, DC, USA, 1971; Volume 105, pp. 126–162. [Google Scholar] [CrossRef]
- Barnett, M.W.; Stenstrom, M.K.; Andrews, J.F. Dynamics and Control of Wastewater Systems, 2nd ed.; CRC Press, Inc.: Lancaster, PA, USA, 1998; Volume 6. [Google Scholar]
- Andrews, J.F. Dynamic models and control strategies for wastewater treatment processes. Water Res. 1974, 8, 261–289. [Google Scholar] [CrossRef]
- Graef, S.P.; Andrews, J.F. Stability and control of anaerobic digestion. J. Water Pollut. Control Fed. 1974, 46, 666–683. [Google Scholar]
- Collins, A.S.; Guilliland, B.E. Control of anaerobic digestion process. J. Environ. Eng. Div. 1974, 2, 487–506. [Google Scholar] [CrossRef]
- Simeonov, I.; Momchev, V.; Grancharov, D. Dynamic modelling of mesophilic anaerobic digestion of animal waste. Water Res. 1996, 30, 1087–1094. [Google Scholar] [CrossRef]
- Andrews, J.F. Development of Control Strategies for Waste-Water Treatment Plants. In Instrumentation Control and Automation for Waste-Water Treatment Systems; Andrews, J., Briggs, R., Jenkins, S., Eds.; Pergamon: New York, NY, USA, 1974; pp. 233–243. [Google Scholar] [CrossRef]
- Olsson, G.; Nielsen, M.; Yuan, Z.; Lynggaard-Jensen, A.; Steyer, J.P. Instrumentation, Control and Automation in Wastewater Systems; IWA Publishing: London, UK, 2005. [Google Scholar] [CrossRef]
- Hill, D.T.; Barth, C.L. A dynamic model for simulation of animal waste digestion. J. Wat. Pollut. Control Fed. 1977, 10, 2129–2143. [Google Scholar]
- Lyberatos, G.; Skiadas, I.V. Modelling of Anaerobic Digestion—A Review. Glob. Nest Int. J. 1999, 1, 63–76. [Google Scholar] [CrossRef]
- Ficara, E.; Hassam, S.; Allegrini, A.; Leva, A.; Malpei, F.; Ferretti, G. Anaerobic Digestion Models: A Comparative Study. IFAC Proc. Vol. 2012, 45, 1052–1057. [Google Scholar] [CrossRef]
- Kythreotou, N.; Florides, G.; Tassou, S.A. A review of simple to scientific models for anaerobic digestion. Renew. Energy 2014, 71, 701–714. [Google Scholar] [CrossRef]
- Hill, D.T. A comprehensive dynamic model for animal waste methanogenesis. Trans. ASAE 1982, 25, 1374–1380. [Google Scholar] [CrossRef]
- Hill, D.T. Design parameters and operating characteristics of animal waste anaerobic digestion systems—Swine and poultry. Agric. Wastes 1983, 5, 157–178. [Google Scholar] [CrossRef]
- Mosey, F.E. Mathematical Modelling of the Anaerobic Digestion Process: Regulatory Mechanisms for the Formation of Short-Chain Volatile Acids from Glucose. Water Sci. Technol. 1983, 15, 209–232. [Google Scholar] [CrossRef]
- Bryers, J.D. Structured modelling of the anaerobic digestion of biomass particulates. Biotechnol. Bioeng. 1984, 27, 638–649. [Google Scholar] [CrossRef]
- Pavlostathis, S.G.; Gossett, J.M. A kinetic model for anaerobic digestion of biological sludge. Biotech. Bioeng. 1986, 28, 1519–1530. [Google Scholar] [CrossRef]
- Moletta, R.; Verrier, D.; Albagnac, G. Dynamic modelling of anaerobic digestion. Water Res. 1986, 20, 427–434. [Google Scholar] [CrossRef]
- Angelidaki, I.; Ellegaard, L.; Ahring, B. A mathematical model for dynamic simulation of anaerobic digestion of complex substrates: Focusing on ammonia inhibition. Biotechnol. Bioeng. 1993, 42, 159–166. [Google Scholar] [CrossRef]
- Siegrist, H.; Renggli, D.; Gujer, W. Mathematical modelling of anaerobic mesophilic sewage sludge treatment. Water Sci. Technol. 1993, 27, 25–36. [Google Scholar] [CrossRef]
- Vavilin, V.A.; Vasiliev, V.B.; Ponomarev, A.V.; Rytow, S.V. Simulation model ‘methane’ as a tool for effective biogas production during anaerobic conversion of complex organic matter. Bioresour. Technol. 1994, 48, 1–8. [Google Scholar] [CrossRef]
- Chen, Y.R.; Hashimoto, A.G. Substrate utilization kinetic model for biological treatment process. Biotechnol. Bioeng. 1980, 22, 2081–2095. [Google Scholar] [CrossRef] [PubMed]
- Gujer, W.; Zehnder, A.J.B. Conversion processes in anaerobic digestion. Water Sci. Technol. 1983, 15, 127–167. [Google Scholar] [CrossRef]
- Dolfing, J. Kinetics of methane formation by granular sludge at low substrate concentrations. Appl. Microbiol. Biotech. 1985, 22, 77–81. [Google Scholar] [CrossRef]
- Gavala, H.N.; Angelidaki, I.; Ahring, B.K. Kinetics and modelling of Anaerobic Digestion Process. In Biomethanation I. Advances in Biochemical Engineering/Biotechnology; Ahring, B.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2003; Volume 81, pp. 126–162. [Google Scholar] [CrossRef]
- Kaspar, H.F. Untersuchungen zur Kopplung von Wasserstoff- und Methanbildung im Faulschlamm [Studies on the Coupling of Hydrogen and Methane Formation in Digested Sludge]. Ph.D. Thesis, Swiss Federal Institute of Technology, Zürich, Switzerland, 1977. [Google Scholar] [CrossRef]
- Irvine, R.L.; Alleman, J.E.; Miller, G.; Dennis, R.W. Stoichiometry and Kinetics of Biological Waste Treatment. J. Water Pollut. Control Fed. 1980, 52, 1997–2006. [Google Scholar] [CrossRef]
- Kleerebezem, R.; van Loosdrecht, M.C.M. Critical analysis of some concepts proposed in ADM1. Water Sci. Technol. 2006, 54, 51–57. [Google Scholar] [CrossRef]
- Petersen, E.E. Chemical Reaction Analysis; Prentice-Hall: Englegood Cliffs, NJ, USA, 1965. [Google Scholar]
- Christ, O.; Wilderer, P.A.; Angerhöfer, R.; Faulstich, M. Mathematical modelling of the hydrolysis of anaerobic processes. Water Sci. Technol. 2000, 41, 61–65. [Google Scholar] [CrossRef]
- Jeyaseelan, S. A simple mathematical model for anaerobic digestion process. Water Sci. Technol. 1997, 35, 185–191. [Google Scholar] [CrossRef]
- Angelidaki, I.; Ellegaard, L.; Ahring, B. A comprehensive model of anaerobic bioconversion of complex substrates to biogas. Biotechnol. Bioeng. 1999, 63, 363–372. [Google Scholar] [CrossRef]
- Blumensaat, F.; Keller, J. Modelling of two-stage anaerobic digestion using the IWA Anaerobic Digestion Model No. 1 (ADM1). Water Res. 2005, 39, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Galí, A.; Benabdallah, T.; Astals, S.; Mata-Alvarez, J. Modified version of ADM1 model for agro-waste application. Bioresour. Technol. 2009, 100, 2783–2790. [Google Scholar] [CrossRef]
- Gaida, D.; Wolf, C.; Meyer, C.; Stuhlsatz, A.; Uppel, J.; Back, T.; Bongards, M.; McLoone, S. State estimation for anaerobic digesters using the ADM1. Water Sci. Technol. 2012, 66, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Liotta, F.; Chatellier, P.; Esposito, G.; Fabbricino, M.; Frunzo, L.; van Hullebusch, E.D.; Lens, P.N.L.; Pirozzi, F. Modified Anaerobic Digestion Model No.1 for dry and semi-dry anaerobic digestion of solid organic waste. Environ. Technol. 2015, 36, 870–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Li, L.; Wu, D.; Xiao, T.; Ma, Y.; Peng, X. Modified Anaerobic Digestion Model No. 1 for modelling methane production from food waste in batch and semi-continuous anaerobic digestions. Bioresour. Technol. 2019, 271, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Batstone, D.J.; Keller, J. Industrial applications of the IWA anaerobic digestion model No. 1 (ADM1). Water Sci. Technol. 2003, 47, 199–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Yang, Z.; Liu, G.; Ma, Z.; Wang, W. Modified anaerobic digestion model No.1 (ADM 1) for modelling anaerobic digestion process at different ammonium concentrations. Water Environ. Res. 2019, 91, 700–714. [Google Scholar] [CrossRef]
- Lauwers, J.; Nimmegeers, P.; Logist, F.; Van Impe, J. Structural identifiability analysis of the Anaerobic Digestion Model No. 1 using a local algebraic observability approach. IFAC-PapersOnLine 2015, 48, 470–475. [Google Scholar] [CrossRef]
- Bernard, O.; Hadj-Sadok, Z.; Dochain, D.; Genovesi, A.; Steyer, J. Dynamical model development and parameter identification for an anaerobic wastewater treatment process. Biotechnol. Bioeng. 2001, 75, 424–438. [Google Scholar] [CrossRef]
- Sbarciog, M.; Loccufier, M.; Noldus, E. Determination of appropriate operating strategies for anaerobic digestion systems. J. Math. Biosci. 2010, 51, 180–188. [Google Scholar] [CrossRef]
- Benyahia, B.; Sari, T.; Cherki, B.; Harmand, J. Bifurcation and stability analysis of a two step model for monitoring anaerobic digestion processes. J. Proc. Control 2012, 22, 1008–1019. [Google Scholar] [CrossRef] [Green Version]
- Sari, T.; Benyahia, B. The operating diagram for a two-step anaerobic digestion model. arXiv 2020, arXiv:math.DS/2004.14028. [Google Scholar]
- Donoso-Bravo, A.; Mailier, J.; Martin, C.; Rodríguez, J.; Arturo Aceves-Lara, C.; Van de Wouwer, A. Model selection, identification and validation in anaerobic digestion: A review. Water Res. 2011, 45, 5347–5364. [Google Scholar] [CrossRef] [PubMed]
- Oreskes, N.; Belitz, K. Philosophical issues in model assessment. In Model Validation: Perspectives in Hydrological Science; Anderson, M.G., Bates, P.D., Eds.; Wiley: Chichester, UK, 2001; pp. 23–41. [Google Scholar]
- Song, H.; Cannon, W.R.; Beliaev, A.S.; Konopka, A. Mathematical modelling of Microbial Community Dynamics: A Methodological Review. Processes 2014, 2, 711–752. [Google Scholar] [CrossRef] [Green Version]
- Laureni, M.; Weissbrodt, D.G.; Villez, K.; Robin, O.; de Jonge, N.; Rosenthal, A.; Wells, G.; Lund Nielsen, J.; Morgenroth, E.; Joss, A. Biomass segregation between biofilm and flocs improves the control of nitrite-oxidizing bacteria in mainstream partial nitritation and anammox processes. Water Res. 2019, 154, 114–116. [Google Scholar] [CrossRef]
- Eberl, H.; Wade, M.J. Challenges and perspectives in reactor scale modelling of biofilm processes. In Recent Trends in Biofilm Science and Technology; Simoes, M., Borges, A., Chaves Simoes, L., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 81–120. [Google Scholar]
- Bernard, O.; Chachuat, B.; Héllias, A.; Rodriguez, J. Can we assess the model complexity for a bioprocess: Theory and example of the anaerobic digestion process. Water Sci. Technol. 2006, 53, 175–182. [Google Scholar] [CrossRef]
- Azeiteiro, C.; Capela, I.F.; Durate, A.C. Dynamic model simulations as a tool for evaluating the stability of an anaerobic process. Water SA 2001, 27, 109–114. [Google Scholar] [CrossRef] [Green Version]
- Steyer, J.P.; Buffiere, P.; Rolland, D.; Moletta, R. Advanced control of anaerobic digestion process through disturbances monitoring. Water Res. 1999, 33, 2059–2068. [Google Scholar] [CrossRef]
- Marsili-Libelli, S.; Nardini, M. Stability and sensitivity analysis of anaerobic digestion models. Environ. Technol. Lett. 1985, 6, 602–609. [Google Scholar] [CrossRef]
- Marsili-Libelli, S.; Beni, S. Shock load modelling in the anaerobic digestion process. Ecol. Modell. 1996, 84, 215–232. [Google Scholar] [CrossRef]
- Brodland, G.W. How computational models can help unlock biological systems. Semin. Cell Dev. Biol. 2015, 47–48, 62–73. [Google Scholar] [CrossRef] [Green Version]
- Suh, N.P. On functional periodicity as the basis for long-term stability of engineered and natural systems and its relationship to physical laws. Res. Eng. Des. 2004, 15, 72–75. [Google Scholar] [CrossRef]
- Bernard, O.; Polit, M.; Hadj-Sadok, Z.; Pengov, M.; Dochain, D.; Estaben, M.; Labat, P. Advanced monitoring and control of anaerobic wastewater treatment plants: Software sensors and controllers for an anaerobic digester. Water Sci. Technol. 2001, 43, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Rincón, A.; Angulo, F.O.G. Control of an anaerobic digester through normal form of fold bifurcation. J. Proc. Cont. 2009, 19, 1355–1367. [Google Scholar] [CrossRef]
- Dimitrova, N.; Krastanov, M. Model-based control strategies for anaerobic digestion processes. Biomath 2019, 8, 1907127. [Google Scholar] [CrossRef] [Green Version]
- Hess, J.; Bernard, O. Design and study of a risk management criterion for an unstable wastewater treatment process. J. Process Control 2008, 18, 71–79. [Google Scholar] [CrossRef]
- Abdelkader, O.H.; Abdelkader, A.H. Modelling Anaerobic Digestion Using Stochastic Approaches. In Trends in Biomathematics: Mathematical modelling for Health, Harvesting, and Population Dynamics: Selected Works Presented at the BIOMAT Consortium Lectures, Morocco 2018; Springer International Publishing: Cham, Switzerland, 2019; pp. 373–396. [Google Scholar] [CrossRef]
- Bailey, J.E. Mathematical modelling and analysis in biochemical engineering: Past accomplishments and future opportunities. Biotechnol. Progress 1998, 14, 8–20. [Google Scholar] [CrossRef]
- Powell, G. Stable coexistence of syntrophic associations in continuous culture. J. Chem. Technol. Biotechnol. 1985, 35, 46–50. [Google Scholar] [CrossRef]
- Hajji, M.; Mazenc, F.; Harmand, J. A mathematical study of syntrophic relationship of a model of anaerobic digestion process. Biosci. Eng. 2010, 7, 641–656. [Google Scholar] [CrossRef]
- Volcke, E.I.P.; Sbarciog, M.; Noldus, E.J.L.; De Baets, B.; Loccufier, M. Steady state multiplicity of two-step biological conversion systems with general kinetics. Math. Biosci. 2010, 228, 160–170. [Google Scholar] [CrossRef] [Green Version]
- Volcke, E.I.P.; Loccufier, M.; Vanrolleghem, P.A.; Noldus, E.J.L. Existence, uniqueness and stability of the equilibrium points of a SHARON bioreactor model. J. Proc. Cont. 2006, 16, 1003–1012. [Google Scholar] [CrossRef]
- Wade, M.J.; Wolkowicz, G.S.K. Bifurcation analysis of an impulsive system describing a Partial Nitritation and Anammox in a hybrid reactor. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wade, M.J.; Pattinson, R.W.; Parker, N.G.; Dolfing, J. Emergent behaviour in a chlorophenol-mineralising three-tiered microbial ‘food web’. J. Theor. Biol. 2016, 389, 171–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sari, T.; Wade, M. Generalised approach to modelling a three-tiered microbial food-web. J. Math. Biosci. 2017, 291, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Premier, G.C.; Guwy, A.; Dinsdale, R. Bifurcation and stability analysis of an anaerobic digestion model. Nonlinear Dynam. 2007, 48, 391–408. [Google Scholar] [CrossRef]
- Rincón, A.; Villa, J.; Angulo, F.O.G.; Olivar, G. A Dynamic Analysis for an Anaerobic Digester: Stability and Bifurcation Branches. Math. Probl. Eng. 2014, 2014, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Bornhöft, A.; Hanke-Rauschenbach, R.; Sundmacher, K. Steady-state analysis of the Anaerobic Digestion Model No. 1 (ADM1). Nonlinear Dynam. 2013, 73, 535–549. [Google Scholar] [CrossRef]
- Weedermann, M.; Seo, G.; Wolkowicz, G.S.K. Mathematical model of anaerobic digestion in a chemostat: Effects of syntrophy and inhibition. J. Biol. Dynam. 2013, 7, 59–85. [Google Scholar] [CrossRef]
- Weedermann, M.; Wolkowicz, G.S.K.; Sasara, J. Optimal biogas production in a model for anaerobic digestion. Nonlinear Dynam. 2015, 81, 1097–1112. [Google Scholar] [CrossRef]
- El-Fadel, M.; Saikaly, P.; Ghanimeh, S. Startup and Stability of Thermophilic Anaerobic Digestion of OFMSW. Crit. Rev. Environ. Sci. Technol. 2013, 43, 2685–2721. [Google Scholar] [CrossRef]
- Treu, L.; Campanaro, S.; Kougias, P.G.; Zhu, X.; Angelidaki, I. Untangling the effect of fatty acids addition at species level revealed different transcriptional responses of the biogas microbial community members. Environ. Sci. Technol. 2016, 50, 6079–6090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campanaro, S.; Treu, L.; Rodriguez-R, L.M.; Kovalovszki, A.; Ziels, R.M.; Maus, I.; Zhu, X.; Kougias, P.G.; Basile, A.; Luo, G.; et al. New insights from the biogas microbiome by comprehensive genome-resolved metagenomics of nearly 1600 species originating from multiple anaerobic digesters. Biotechnol. Biofuels 2020, 13, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardona, L.; Lê Cao, K.A.; Puig-Castellví, F.; Bureau, C.; Madigou, C.; Mazéas, L.; Chapleur, O. Integrative analyses to investigate the link between microbial activity and metabolites degradation during anaerobic digestion. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Hassa, J.; Maus, I.; Off, S.; Pühler, A.; Scherer, P.; Klocke, M.; Schlüter, A. Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants. Appl. Microbiol. Biotechnol. 2018, 102, 5045–5063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X. Deciphering the Microbial Ecology in Biogas Reactors for Optimizing the Anaerobic Digestion Process. Ph.D. Thesis, Department of Environmental Engineering, Technical University of Denmark (DTU), Lyngby, Denmark, 2018. [Google Scholar]
- Poirier, S.; Déjean, S.; Midoux, C.; Lê Cao, K.; Chapleur, O. Integrating independent microbial studies to build predictive models of anaerobic digestion inhibition. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Gimpel, G. Bringing dark data into the light: Illuminating existing IoT data lost within your organization. Bus. Horiz. 2020, 63, 519–530. [Google Scholar] [CrossRef]
- Wade, M.J. The Digital Frontier: A Perspective on Digitalisation for Water. Inst. Water Mag. 2020, 205, 30–31. [Google Scholar]
- Madsen, M.; Holm-Nielsen, J.B.; Esbensen, K.H. Monitoring of anaerobic digestion processes: A review perspective. Renew. Sust. Energ. Rev. 2011, 15, 3141–3155. [Google Scholar] [CrossRef] [Green Version]
- Kazemi, P.; Giralt, J.; Bengoa, C.; Steyer, J. Data-driven fault detection methods for detecting small-magnitude faults in anaerobic digestion process. Water Sci. Technol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Jimenez, J.; Latrille, E.; Harmand, J.; Robles, Á.; Ferrer, J.; Gaida, D.; Wolf, C.; Mairet, F.; Bernard, O.; Alcaraz-González, V.; et al. Instrumentation and control of anaerobic digestion processes: A review and some research challenges. Rev. Environ. Sci. Biotechnol. 2015, 14, 615–648. [Google Scholar] [CrossRef]
- Asadi, M.; Guo, H.; McPhedran, K. Biogas production estimation using data-driven approaches for cold region municipal wastewater anaerobic digestion. J. Environ. Manag. 2020, 253, 109708. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.P.; Devlin, D.; Esteves, S.R.; Dinsdale, R.; Guwy, A.J. Integration of NIRS and PCA techniques for the process monitoring of a sewage sludge anaerobic digester. Bioresour. Technol. 2013, 133, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Haimi, H.; Mulas, M.; Corona, F.; Vahala, R. Data-derived soft-sensors for biological wastewater treatment plants: An overview. Environ. Model. Softw. 2013, 47, 88–107. [Google Scholar] [CrossRef]
- Lee, D.; Vanrolleghem, P.A. Adaptive Consensus Principal Component Analysis for On-Line Batch Process Monitoring. Environ. Monit. Assess. 2004, 92, 119–135. [Google Scholar] [CrossRef] [Green Version]
- Wade, M.J.; Sánchez, A.; Katebi, M.R. On real-time control and process monitoring of wastewater treatment plants: Real-time process monitoring. Trans. Inst. Meas. Cont. 2005, 27, 173–193. [Google Scholar] [CrossRef]
- Lemaigre, S.; Adam, G.; Gerin, P.A.; Noo, A.; De Vos, B.; Klimek, D.; Goux, X.; Calusinska, M.; Delfosse, P. Potential of multivariate statistical process monitoring based on the biogas composition to detect free ammonia intoxication in anaerobic reactors. Biochem. Eng. J. 2018, 140, 17–28. [Google Scholar] [CrossRef]
- Corominas, L.; Villez, K.; Aguado, D.; Rieger, L.; Rosén, C.; Vanrolleghem, P.A. Performance evaluation of fault detection methods for wastewater treatment processes. Biotechnol. Bioeng. 2011, 108, 333–344. [Google Scholar] [CrossRef]
- Corominas, L.; Garrido-Baserba, M.; Villez, K.; Olsson, G.; Cortés, U.; Poch, M. Transforming data into knowledge for improved wastewater treatment operation: A critical review of techniques. Environ. Model. Softw. 2018, 106, 89–103. [Google Scholar] [CrossRef]
- Oppong, G.; O’Brien, M.; McEwan, M.; Martin, E.B.; Montague, G.A. Advanced Control for Anaerobic Digestion Processes: Volatile Solids Soft Sensor Development. Comput. Aided Chem. Eng. 2012, 30, 967–971. [Google Scholar] [CrossRef]
- Awhangbo, L.; Bendoula, R.; Roger, J.M.; Béline, F. Multi-block SO-PLS approach based on infrared spectroscopy for anaerobic digestion process monitoring. Chemometr. Intell. Lab. Syst. 2020, 196, 103905. [Google Scholar] [CrossRef]
- Premier, G.C.; Dinsdale, R.; Guwy, A.J.; Hawkes, F.R.; Hawkes, D.L.; Wilcox, S.J. A comparison of the ability of black box and neural network models of ARX structure to represent a fluidized bed anaerobic digestion process. Water Res. 1999, 33, 1027–1037. [Google Scholar] [CrossRef]
- Simeonov, I.; Chorukova, E.; Kalchev, B. Anaerobic Digestion Modelling with Artificial Neural Networks. IFAC Proc. Vol. 2004, 37, 225–230. [Google Scholar] [CrossRef]
- Turkdogan-Aydınol, F.I.; Yetilmezsoy, K. A fuzzy-logic-based model to predict biogas and methane production rates in a pilot-scale mesophilic UASB reactor treating molasses wastewater. J. Hazard. Mater. 2010, 182, 460–471. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, D.; Wen, Z.G.; Fei, F.; Caicedo, L.; Yuan, K.; Shang, R.X. Interpretable machine learning for predicting biomethane production in industrial-scale anaerobic co-digestion. Sci. Total Environ. 2020, 712, 134574. [Google Scholar] [CrossRef]
- Tufaner, F.; Demirci, Y. Prediction of biogas production rate from anaerobic hybrid reactor by artificial neural network and nonlinear regressions models. Clean Technol. Environ. Policy 2020, 22, 713–724. [Google Scholar] [CrossRef]
- Baker, R.E.; Na, J.P.; Jayamohan, J.; Jérusalem, A. Mechanistic models versus machine learning, a fight worth fighting for the biological community? Biol. Lett. 2018, 14, 20170660. [Google Scholar] [CrossRef]
- Karama, A.; Bernard, O.; Gouzé, J.L.; Benhammou, A.; Dochain, D. Hybrid neural modelling of an anaerobic digester with respect to biological constraints. Water Sci. Technol. 2001, 43, 1–8. [Google Scholar] [CrossRef]
- Lopez, P.C.; Udugama, I.A.; Thomsen, S.T.; Roslander, C.; Junicke, H.; Mauricio-Iglesias, M.; Gernaey, K.V. Towards a digital twin: A hybrid data-driven and mechanistic digital shadow to forecast the evolution of lignocellulosic fermentation. Biofuels Bioprod. Biorefin. 2020. [Google Scholar] [CrossRef]
- Kusiak, A.; Wei, X. A data-driven model for maximization of methane production in a wastewater treatment plant. Water Sci. Technol. 2012, 65, 1116–1122. [Google Scholar] [CrossRef]
- Ramachandran, A.; Rustum, R.; Adeloye, A.J. Review of Anaerobic Digestion modelling and Optimization Using Nature-Inspired Techniques. Processes 2019, 7, 953. [Google Scholar] [CrossRef] [Green Version]
- García-Diéguez, C.; Bernard, O.; Roca, E. Reducing the Anaerobic Digestion Model No. 1 for its application to an industrial wastewater treatment plant treating winery effluent wastewater. Bioresour. Technol. 2013, 132, 244–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinrich, S.; Koch, S.; Bonk, F.; Popp, D.; Benndorf, D.; Klamt, S.; Centler, F. Augmenting Biogas Process modelling by Resolving Intracellular Metabolic Activity. Front. Microbiol. 2019, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
- Großkopf, T.; Soyer, O.S. Microbial diversity arising from thermodynamic constraints. ISME J. 2016, 10, 2725–2733. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Yang, P.; Singh, S.; Zhuang, H.; Xu, L.; Chen, W.; Dolfing, J.; Li, D.; Zhang, Y.; Zeng, H.; et al. A review on the bioenergetics of anaerobic microbial metabolism close to the thermodynamic limits and its implications for digestion applications. Bioresour. Technol. 2018, 247, 1095–1106. [Google Scholar] [CrossRef]
- Kleerebezem, R.; van Loosdrecht, M.C.M. A Generalized Method for Thermodynamic State Analysis of Environmental Systems. Crit. Rev. Environ. Sci. Technol. 2010, 40, 1–54. [Google Scholar] [CrossRef]
- McCarty, P.L. Thermodynamics of biological synthesis and growth. Air Water Pollut. 1965, 9, 621–639. [Google Scholar]
- McCarty, P.L. Energetics and Kinetics of Anaerobic Treatment. In Anaerobic Biological Treatment Processes; American Chemical Society: Washington, DC, USA, 1971; Volume 105, pp. 91–107. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, Y.M.; Yang, S.F. A thermodynamic interpretation of the Monod equation. Curr. Microbiol. 2003, 46, 233–234. [Google Scholar] [CrossRef]
- Hoh, C.Y.; Cord-Ruwisch, R. A practical kinetic model that considers end-product inhibition in anaerobic digestion processes by including the equilibrium constant. Biotechnol. Bioeng. 1996, 51, 597–604. [Google Scholar] [CrossRef]
- Haldane, J.B.S. Enzymes; Monographs on Biochemistry; Longmans, Green: London, UK, 1930. [Google Scholar]
- Atkins, P.; de Paula, J.; Keeler, J. Physical Chemistry, 11th ed.; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Jin, Q.; Bethke, C.M. A new rate law describing microbial respiration. Appl. Environ. Microbiol. 2003, 69, 2340–2348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, J.; Lema, J.M.; Kleerebezem, R. Energy-based models for environmental biotechnology. Trends Biotechnol. 2008, 26, 366–374. [Google Scholar] [CrossRef]
- González-Cabaleiro, R.; Lema, J.M.; Rodríguez, J.; Kleerebezum, R. Linking thermodynamics and kinetics to assess pathway reversibility in anaerobic bioprocesses. Energy Environ. Sci. 2013, 6, 3780–3789. [Google Scholar] [CrossRef]
- Jin, Q.; Bethke, C.M. The thermodynamics and kinetics of microbial metabolism. Am. J. Sci. 2007, 307, 643–677. [Google Scholar] [CrossRef]
- González-Cabaleiro, R.; Ofiţeru, I.D.; Lema, J.M.; Rodríguez, J. Microbial catabolic activities are naturally selected by metabolic energy harvest rate. ISME J. 2015, 9, 2630–2641. [Google Scholar] [CrossRef] [PubMed]
- LaRowe, D.E.; Dale, A.W.; Amend, J.P.; Van Cappellen, P. Thermodynamic limitations on microbially catalyzed reaction rates. Geochim. Cosmochim. Acta 2012, 90, 96–109. [Google Scholar] [CrossRef]
- Desmond-Le Quéméner, E.; Bouchez, T. A thermodynamic theory of microbial growth. ISME J. 2014, 8, 1747–1751. [Google Scholar] [CrossRef] [PubMed]
- Delattre, H.; Desmond-Le Quéméner, E.; Duquennoi, C.; Filali, A.; Bouchez, T. Consistent microbial dynamics and functional community patterns derived from first principles. ISME J. 2019, 13, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.T.; Martin, A.D. Thermodynamic equilibrium model in anaerobic digestion process. Biochem. Eng. J. 2007, 34, 256–266. [Google Scholar] [CrossRef]
- Delattre, H.; Chen, J.; Wade, M.J.; Soyer, O.S. Thermodynamic modelling of synthetic communities predicts minimum free energy requirements for sulfate reduction and methanogenesis. J. R. Soc. Interface 2020, 17, 20200053. [Google Scholar] [CrossRef]
- Gaebler, H.J.; Eberl, H.J. Thermodynamic Inhibition in Chemostat Models: With an Application to Bioreduction of Uranium. Bull. Math. Biol. 2020, 82, 76. [Google Scholar] [CrossRef]
- Moscoviz, R.; Trably, E.; Bernet, N.; Carrère, H. The environmental biorefinery: State-of-the-art on the production of hydrogen and value-added biomolecules in mixed-culture fermentation. Green Chem. 2018, 20, 3159–3179. [Google Scholar] [CrossRef]
- Regueira, A.; Lema, J.; Carballa, M.; Mauricio-Iglesias, M. Metabolic modelling for predicting VFA production from protein-rich substrates by mixed-culture fermentation. Biotechnol. Bioeng. 2020, 117, 73–84. [Google Scholar] [CrossRef]
- Righetti, E.; Nortilli, S.; Fatone, F.; Frison, N.; Bolzonella, D. A Multiproduct Biorefinery Approach for the Production of Hydrogen, Methane and Volatile Fatty Acids from Agricultural Waste. Waste Biomass Valor. 2020. [Google Scholar] [CrossRef] [Green Version]
- Ganidi, N.; Tyrrel1, S.; Cartmell, E. Anaerobic Digestion Foaming Causes—A review. Bioresour. Technol. 2009, 100, 5546–5554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.; Kuo-Dahab, C.; Chapman, T.; Mei, Y. Anaerobic digestion, mixing, environmental fate, and transport. Water Environ. Res. 2019, 91, 1210–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samstag, R.W.; Ducoste, J.J.; Griborio, A.; Nopens, I.; Batstone, D.J.; Wicks, J.D.; Saunders, S.; Wicklein, E.A.; Kenny, G.; Laurent, J. CFD for wastewater treatment: An overview. Water Sci. Technol. 2016, 74, 549–563. [Google Scholar] [CrossRef]
- Sadino-Riquelme, C.; Hayes, R.E.; Jeison, D.; Donoso-Bravo, A. Computational fluid dynamic (CFD) modelling in anaerobic digestion: General application and recent advances. Crit. Rev. Environ. Sci. Technol. 2018, 48, 39–76. [Google Scholar] [CrossRef]
- Dapelo, D.; Alberini, F.; Bridgeman, J. Euler-Lagrange CFD modelling of unconfined gas mixing in anaerobic digestion. Water Res. 2015, 85, 497–511. [Google Scholar] [CrossRef] [Green Version]
- Terashima, M.; Goel, R.; Komatsu, K.; Yasui, H.; Takahashi, H.; Li, Y.Y.; Noike, T. CFD simulation of mixing in anaerobic digesters. Bioresour. Technol. 2009, 100, 2228–2233. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, G.; Yu, L.; Siddhu, M.A.H.; Gao, M.; Abdeltawab, A.A.; Al-Deyab, S.S.; Chen, X. Computational fluid dynamics study on mixing mode and power consumption in anaerobic mono- and co-digestion. Bioresour. Technol. 2016, 203, 166–172. [Google Scholar] [CrossRef]
- Wu, B. Advances in the use of CFD to characterize, design and optimize bioenergy systems. Comp. Electron. Agric. 2013, 93, 195–208. [Google Scholar] [CrossRef]
- Rezavand, M.; Winkler, D.; Sappl, J.; Seiler, L.; Meister, M.; Rauch, W. A fully Lagrangian computational model for the integration of mixing and biochemical reactions in anaerobic digestion. Comput. Fluids 2019, 181, 224–235. [Google Scholar] [CrossRef]
- Tobo, Y.M.; Rehman, U.; Bartacek, J.; Nopens, I. Partial integration of ADM1 into CFD: Understanding the impact of diffusion on anaerobic digestion mixing. Water Sci. Technol. 2020, 81, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Picioreanu, C.; van Loosdrecht, M.C.M.; Heijnen, J.J. Mathematical modelling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol. Bioeng. 1998, 58, 101–116. [Google Scholar] [CrossRef]
- Kreft, J.; Booth, G.; Wimpenny, J.W.T. BacSim, a simulator for individual-based modelling of bacterial colony growth. Microbiology 1998, 144, 3275–3287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picioreanu, C.; van Loosdrecht, M.C.M.; Heijnen, J.J. Discrete-differential modelling of biofilm structure. Water Sci. Technol. 1999, 39, 115–122. [Google Scholar] [CrossRef]
- Ardré, M.; Henry, H.; Douarche, C.; Plapp, M. An individual-based model for biofilm formation at liquid surfaces. Phys. Biol. 2015, 12, 066015. [Google Scholar] [CrossRef] [Green Version]
- García, M.R.; Vázquez, J.A.; Teixeira, I.G.; Alonso, A.A. Stochastic Individual-Based modelling of Bacterial Growth and Division Using Flow Cytometry. Front. Microbiol. 2018, 8, 2626. [Google Scholar] [CrossRef] [Green Version]
- Gogulancea, V.; González-Cabaleiro, R.; Li, B.; Taniguchi, D.; Jayathilake, P.G.; Chen, J.; Wilkinson, D.; Swailes, D.; McGough, A.S.; Zuliani, P.; et al. Individual Based Model Links Thermodynamics, Chemical Speciation and Environmental Conditions to Microbial Growth. Front. Microbiol. 2019, 10, 1871. [Google Scholar] [CrossRef] [Green Version]
- Jayathilake, P.G.; Gupta, P.; Li, B.; Madsen, C.; Oyebamiji, O.; González-Cabaleiro, R.; Rushton, S.; Bridgens, B.; Swailes, D.; Allen, B.; et al. A mechanistic Individual-based Model of microbial communities. PLoS ONE 2017, 12, e0181965. [Google Scholar] [CrossRef] [Green Version]
- Oyebamiji, O.K.; Wilkinson, D.J.; Li, B.; Jayathilake, P.G.; Zuliani, P.; Curtis, T.P. Bayesian emulation and calibration of an individual-based model of microbial communities. J. Comput. Sci. 2019, 30, 194–208. [Google Scholar] [CrossRef]
- Doloman, A.; Varghese, H.; Miller, C.D.; Flann, N.S. modelling de novo granulation of anaerobic sludge. BMC Syst. Biol. 2017, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Taniguchi, D.; Gedara, J.P.; Gogulancea, V.; Gonzalez-Cabaleiro, R.; Chen, J.; McGough, A.S.; Ofiţeru, I.D.; Curtis, T.P.; Zuliani, P. NUFEB: A massively parallel simulator for individual-based modelling of microbial communities. PLoS Comput. Biol. 2019, 15, e1007125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levins, R. Evolution in communities near equilibrium. In Ecology and Evolution of Communities; Cody, M.L., MacArthur, R.H., Diamond, J.M., Eds.; Harvard Univ. Press: Cambridge, MA, USA, 1975; pp. 81–120. [Google Scholar]



| Model | # State Variables | # Parameters | Year | Program | Notes | Reference |
|---|---|---|---|---|---|---|
| [AGa →AMG] | 9 | 28 | 1977 | CSMP | VFA and NH inhibition on MG only | [41] |
| [AGa →AMG + HMG] | 4 | N/A * | 1982 | CSMP | Complete use of H assumed | [45,46] |
| [AGp + AGb →AcG→AMG + HMG] | 9 | 18 | 1983 | NR | pH regulation through redox potential by H | [47] |
| [EH →AG→pAcG→AMG + HMG] | 10 | 35 | 1984 | MIMIC | First generalised and structured model (stoichiometric matrix) | [48] |
| [EH →AGa→AMG] | 7 | 14 | 1986 | NR | Hydrolysis of dead, particulate biomass is rate-limiting step | [49] |
| [AGa →AMG] | 5 | 23 | 1986 | NR | VFA inhibition on AG & MG, NH inhibition on MG | [50] |
| [EH → AG → pAcG + bAcG →AMG] | 16 | 33 | 1993 | Pascal | NH inhibition & pH-Temperature interactions | [51] |
| [EH →AG→pAcG + fAcG→AMG + HMG] | 18 | 48 | 1993 | Pascal | Extension of structured approach. Focus on acetate kinetics/digester stability | [52] |
| [EH →AGp + AGa→AcG + pSR + aSR→AMG + HMG] | 33 | 55 | 1994 | METHANE | H inhibition considered | [53] |
| [EH →AG→MG] | 5 | 14 | 1996 | TUTSIM | Reduced order for process control | [38] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wade, M.J. Not Just Numbers: Mathematical Modelling and Its Contribution to Anaerobic Digestion Processes. Processes 2020, 8, 888. https://0-doi-org.brum.beds.ac.uk/10.3390/pr8080888
Wade MJ. Not Just Numbers: Mathematical Modelling and Its Contribution to Anaerobic Digestion Processes. Processes. 2020; 8(8):888. https://0-doi-org.brum.beds.ac.uk/10.3390/pr8080888
Chicago/Turabian StyleWade, Matthew J. 2020. "Not Just Numbers: Mathematical Modelling and Its Contribution to Anaerobic Digestion Processes" Processes 8, no. 8: 888. https://0-doi-org.brum.beds.ac.uk/10.3390/pr8080888





