The Maturation Pathway of Nickel Urease
Abstract
:1. Introduction
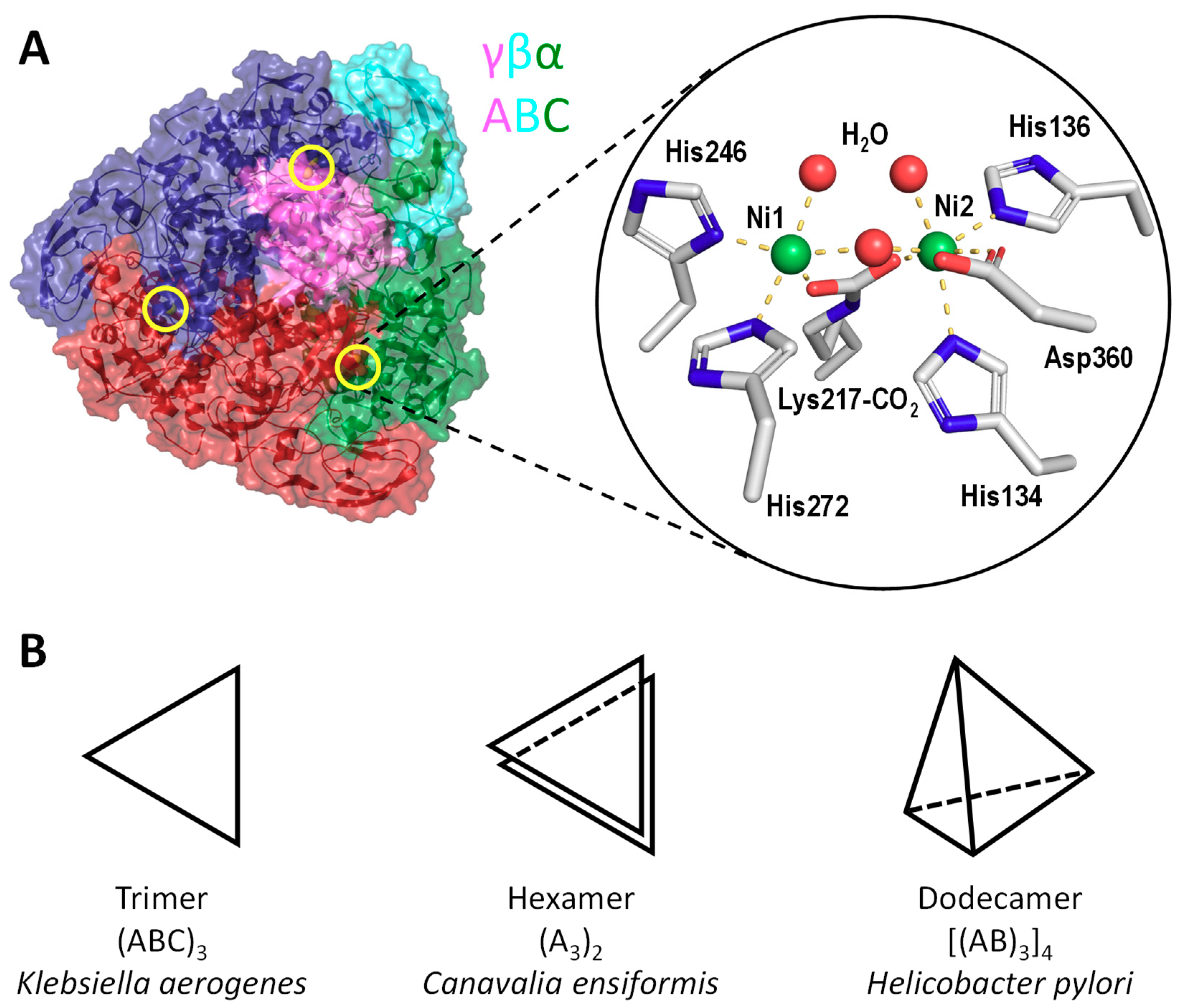
2. Structures of Urease
3. Genetic Studies Showed the Importance of Urease Accessory Proteins
4. The Formation of UreGFD Complex
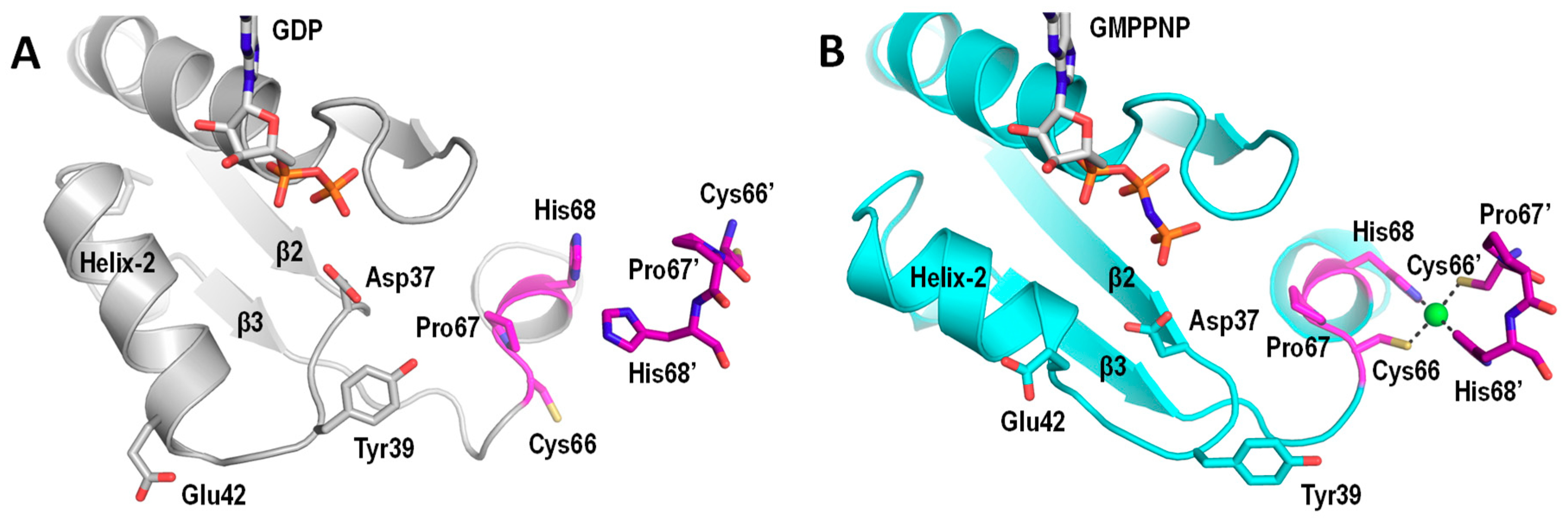
5. UreG Dissociates from the UreGFD Complex and Forms a Dimer in the Presence of Ni/GTP
6. UreG–UreE Interaction Is GTP-Dependent
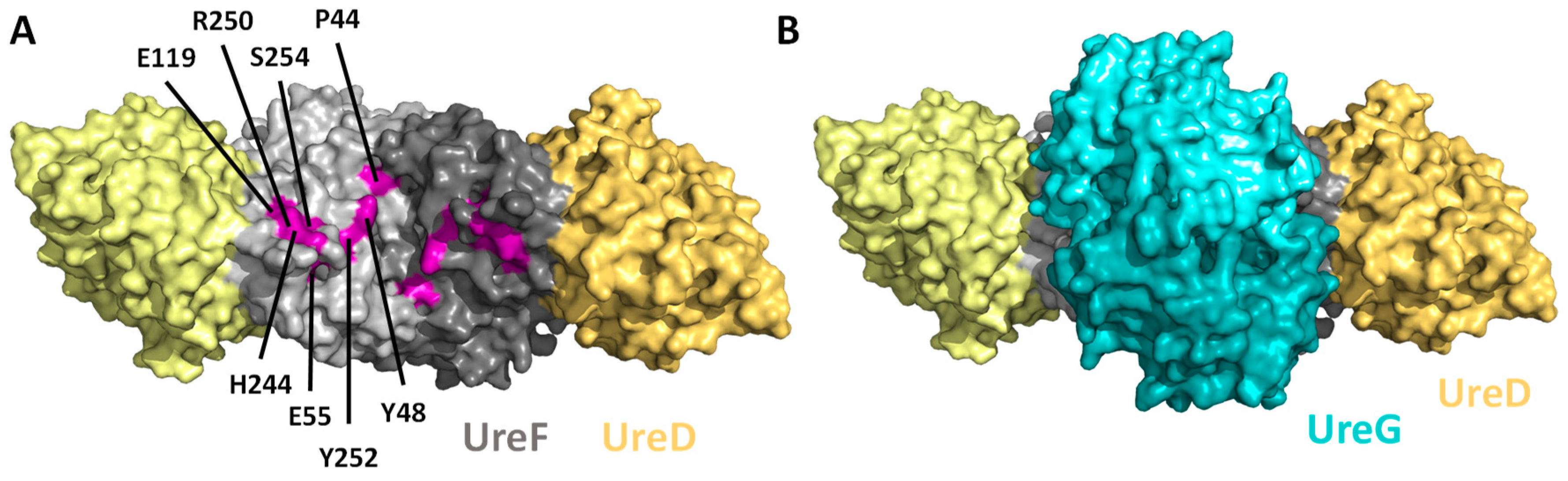
7. How Urease Accessory Proteins Facilitate Urease Maturation
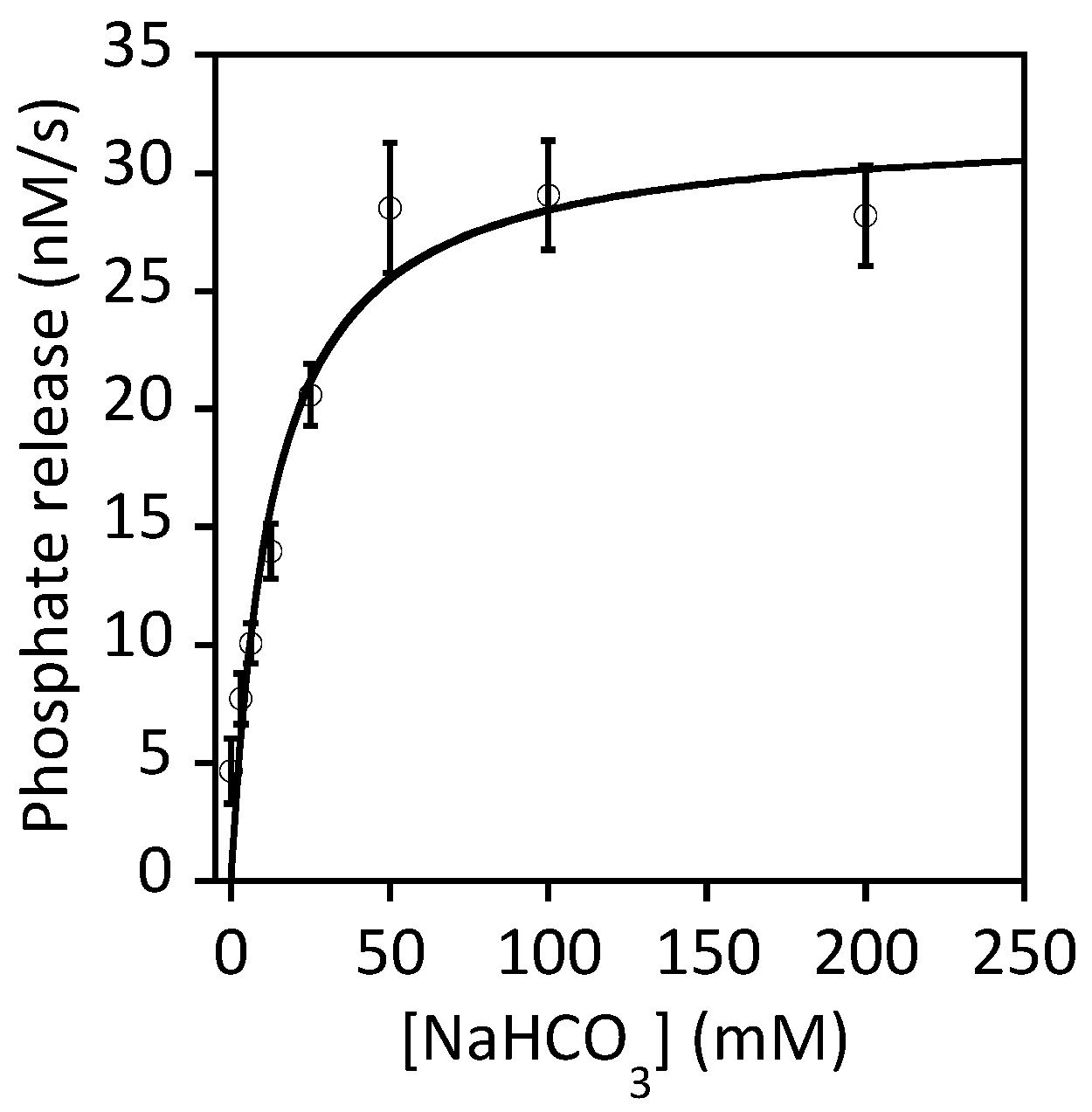
8. UreE Gets Its Nickel from Cross-Talking to the Hydrogenase Maturation Pathway
9. Urease Maturation Pathway in Plants
10. Urease Maturation Pathway Is Druggable
11. Future Perspectives
12. Conclusions
Funding
Conflicts of Interest
References
- Krajewska, B. Ureases I. Functional, Catalytic and Kinetic Properties: A Review. J. Mol. Catal. B Enzym. 2009, 59, 9–21. [Google Scholar] [CrossRef]
- Hanlon, D.P. The Distribution of Arginase and Urease in Marine Invertebrates. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1975, 52, 261–264. [Google Scholar] [CrossRef]
- Alonso-Saez, L.; Waller, A.S.; Mende, D.R.; Bakker, K.; Farnelid, H.; Yager, P.L.; Lovejoy, C.; Tremblay, J.-E.; Potvin, M.; Heinrich, F.; et al. Role for Urea in Nitrification by Polar Marine Archaea. Proc. Natl. Acad. Sci. USA 2012, 109, 17989–17994. [Google Scholar] [CrossRef] [PubMed]
- Dixon, N.E.; Gazzola, C.; Watters, J.J.; Blakeley, R.L.; Zerner, B. Jack Bean Urease (EC 3.5.1.5). A Metalloenzyme. A Simple Biological Role for Nickel? J. Am. Chem. Soc. 1975, 97, 4131–4133. [Google Scholar] [CrossRef] [PubMed]
- Sumner, J.B. The Isolation and Crystallization of the Enzyme Urease. J. Biol. Chem. 1926, 69, 435–442. [Google Scholar]
- Kao, C.-Y.; Sheu, B.-S.; Wu, J.-J. Helicobacter pylori infection: An overview of bacterial virulence facztors and pathogenesis. Biomed. J. 2016, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, S.; Nunn, L. Helicobacter pylori and Gastric Cancer: A State of the Art Review. Gastroenterol. Hepatol. Bed Bench 2015, 8, S6–S14. [Google Scholar] [PubMed]
- Narayanan, M.; Reddy, K.M.; Marsicano, E. Peptic Ulcer Disease and Helicobacter pylori Infection. Mo. Med. 2018, 115, 219–224. [Google Scholar] [PubMed]
- Carter, E.L.; Tronrud, D.E.; Taber, S.R.; Karplus, P.A.; Hausinger, R.P. Iron-Containing Urease in a Pathogenic Bacterium. Proc. Natl. Acad. Sci. USA 2011, 108, 13095–13099. [Google Scholar] [CrossRef]
- Capdevila, D.A.; Edmonds, K.A.; Giedroc, D.P. Metallochaperones and Metalloregulation in Bacteria. Essays Biochem. 2017, 61, 177–200. [Google Scholar] [CrossRef]
- Waldron, K.J.; Robinson, N.J. How Do Bacterial Cells Ensure That Metalloproteins Get the Correct Metal? Nat. Rev. Microbiol. 2009, 7, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Kappaun, K.; Piovesan, A.R.; Carlini, C.R.; Ligabue-Braun, R. Ureases: Historical Aspects, Catalytic, and Non-Catalytic Properties—A Review. J. Adv. Res. 2018, 13, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Todd, M.J.; Hausinger, R.P. Purification and Characterization of the Nickel-Containing Multicomponent Urease from Klebsiella aerogenes. J. Biol. Chem. 1987, 262, 5963–5967. [Google Scholar] [PubMed]
- Hu, L.T.; Mobley, H.L. Purification and N-Terminal Analysis of Urease from Helicobacter pylori. Infect. Immun. 1990, 58, 992–998. [Google Scholar] [PubMed]
- Blakeley, R.L.; Zerner, B. Jack Bean Urease: The First Nickel Enzyme. J. Mol. Catal. 1984, 23, 263–292. [Google Scholar] [CrossRef]
- Pearson, M.A.; Michel, L.O.; Hausinger, R.P.; Karplus, P.A. Structures of Cys319 Variants and Acetohydroxamate-Inhibited Klebsiella aerogenes Urease. Biochemistry 1997, 36, 8164–8172. [Google Scholar] [CrossRef] [PubMed]
- Jabri, E.; Carr, M.B.; Hausinger, R.P.; Karplus, P.A. The Crystal Structure of Urease from Klebsiella aerogenes. Science 1995, 268, 998–1004. [Google Scholar] [CrossRef]
- Benini, S.; Rypniewski, W.R.; Wilson, K.S.; Miletti, S.; Ciurli, S.; Mangani, S. A New Proposal for Urease Mechanism Based on the Crystal Structures of the Native and Inhibited Enzyme from Bacillus pasteurii: Why Urea Hydrolysis Costs Two Nickels. Structure 1999, 7, 205–216. [Google Scholar] [CrossRef]
- Balasubramanian, A.; Ponnuraj, K. Crystal Structure of the First Plant Urease from Jack Bean: 83 Years of Journey from Its First Crystal to Molecular Structure. J. Mol. Biol. 2010, 400, 274–283. [Google Scholar] [CrossRef]
- Ha, N.; Oh, S.; Sung, J.Y.; Cha, K.A.; Lee, M.H.; Oh, B. Supramolecular Assembly and Acid Resistance of Helicobacter pylori Urease. Nat. Struct. Mol. Biol. 2001, 8, 505–509. [Google Scholar] [CrossRef]
- Ciurli, S.; Benini, S.; Rypniewski, W.R.; Wilson, K.S.; Miletti, S.; Mangani, S. Structural Properties of the Nickel Ions in Urease: Novel Insights into the Catalytic and Inhibition Mechanisms. Coord. Chem. Rev. 1999, 190–192, 331–355. [Google Scholar] [CrossRef]
- Park, I.S.; Hausinger, R.P. Requirement of Carbon Dioxide for in vitro Assembly of the Urease Nickel Metallocenter. Science 1995, 267, 1156–1158. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.A.; Schaller, R.A.; Michel, L.O.; Karplus, P.A.; Hausinger, R.P. Chemical Rescue of Klebsiella aerogenes Urease Variants Lacking the Carbamylated-Lysine Nickel Ligand. Biochemistry 1998, 37, 6214–6220. [Google Scholar] [CrossRef] [PubMed]
- Park, I.S.; Hausinger, R.P. Metal Ion Interactions with Urease and UreD-Urease Apoproteins. Biochemistry 1996, 35, 5345–5352. [Google Scholar] [CrossRef] [PubMed]
- Mulrooney, S.B.; Hausinger, R.P. Sequence of the Klebsiella aerogenes Urease Genes and Evidence for Accessory Proteins Facilitating Nickel Incorporation. J. Bacteriol. 1990, 172, 5837–5843. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Mulrooney, S.B.; Renner, M.J.; Markowicz, Y.; Hausinger, R.P. Klebsiella aerogenes Urease Gene Cluster: Sequence of UreD and Demonstration That Four Accessory Genes (UreD, UreE, UreF, and UreG) Are Involved in Nickel Metallocenter Biosynthesis. J. Bacteriol. 1992, 174, 4324–4330. [Google Scholar] [CrossRef] [PubMed]
- Akada, J.K.; Shirai, M.; Takeuchi, H.; Tsuda, M.; Nakazawa, T. Identification of the Urease Operon in Helicobacter pylori and Its Control by mRNA Decay in Response to pH. Mol. Microbiol. 2000, 36, 1071–1084. [Google Scholar] [CrossRef]
- Cussac, V.; Ferrero, R.L.; Labigne, A. Expression of Helicobacter pylori Urease Genes in Escherichia coli Grown under Nitrogen-Limiting Conditions. J. Bacteriol. 1992, 174, 2466–2473. [Google Scholar] [CrossRef]
- Weeks, D.L.; Eskandari, S.; Scott, D.R.; Sachs, G. A H+-Gated Urea Channel: The Link between Helicobacter pylori Urease and Gastric Colonization. Science 2000, 287, 482–485. [Google Scholar] [CrossRef]
- Rektorschek, M.; Buhmann, A.; Weeks, D.; Schwan, D.; Bensch, K.W.; Eskandari, S.; Scott, D.; Sachs, G.; Melchers, K. Acid Resistance of Helicobacter pylori Depends on the UreI Membrane Protein and an Inner Membrane Proton Barrier. Mol. Microbiol. 2000, 36, 141–152. [Google Scholar] [CrossRef]
- Benoit, S.L.; Zbell, A.L.; Maier, R.J. Nickel Enzyme Maturation in Helicobacter hepaticus: Roles of Accessory Proteins in Hydrogenase and Urease Activities. Microbiology 2007, 153, 3748–3756. [Google Scholar] [CrossRef] [PubMed]
- Sriwanthana, B.; Island, M.D.; Maneval, D.; Mobley, H.L.T. Single-Step Purification of Proteus mirabilis Urease Accessory Protein UreE, a Protein with a Naturally Occurring Histidine Tail, by Nickel Chelate Affinity Chromatography. J. Bacteriol. 1994, 176, 6836–6841. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Foxall, P.A.; Russell, R.; Mobley, H.L.T. Purification of Recombinant Helicobacter pylori Urease Apoenzyme Encoded by UreA and UreB. Infect. Immun. 1992, 60, 2657–2667. [Google Scholar] [PubMed]
- Lee, M.H.; Mulrooney, S.B.; Hausinger, R.P. Purification, Characterization, and in vivo Reconstitution of Klebsiella aerogenes Urease Apoenzyme. J. Bacteriol. 1990, 172, 4427–4431. [Google Scholar] [CrossRef] [PubMed]
- Voland, P.; Weeks, D.L.; Marcus, E.A.; Prinz, C.; Sachs, G.; Scott, D. Interactions among the Seven Helicobacter pylori Proteins Encoded by the Urease Gene Cluster. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 284, G96–G106. [Google Scholar] [CrossRef] [PubMed]
- Benoit, S.L.; Mehta, N.; Weinberg, M.V.; Maier, C.; Maier, R.J. Interaction between the Helicobacter pylori Accessory Proteins HypA and UreE Is Needed for Urease Maturation. Microbiology 2007, 153, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
- Park, I.S.; Carr, M.B.; Hausinger, R.P. In vitro Activation of Urease Apoprotein and Role of UreD as a Chaperone Required for Nickel Metallocenter Assembly. Proc. Natl. Acad. Sci. USA 1994, 91, 3233–3237. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.L.; Hausinger, R.P. Characterization of the Klebsiella aerogenes Urease Accessory Protein UreD in Fusion with the Maltose Binding Protein. J. Bacteriol. 2010, 192, 2294–2304. [Google Scholar] [CrossRef]
- Moncrief, M.B.C.; Hausinger, R.P. Purification and Activation Properties of UreD-UreF-Urease Apoprotein Complexes. J. Bacteriol. 1996, 178, 5417–5421. [Google Scholar] [CrossRef]
- Kim, J.K.; Mulrooney, S.B.; Hausinger, R.P. The UreEF Fusion Protein Provides a Soluble and Functional Form of the UreF Urease Accessory Protein. J. Bacteriol. 2006, 188, 8413–8420. [Google Scholar] [CrossRef] [Green Version]
- Soriano, A.; Hausinger, R.P. GTP-Dependent Activation of Urease Apoprotein in Complex with the UreD, UreF, and UreG Accessory Proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 11140–11144. [Google Scholar] [CrossRef] [PubMed]
- Park, I.; Hausinger, R.P. Evidence for the Presence of Urease Apoprotein Complexes Containing UreD, UreF, and UreG in Cells That Are Competent for in vivo Enzyme Activation. J. Bacteriol. 1995, 177, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Fong, Y.H.; Wong, H.C.; Yuen, M.H.; Lau, P.H.; Chen, Y.W.; Wong, K.-B. Structure of UreG/UreF/UreH Complex Reveals How Urease Accessory Proteins Facilitate Maturation of Helicobacter pylori Urease. PLoS Biol. 2013, 11, e1001678. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, M.A.; Han, L.; Zhong, Y.; Boer, J.L.; Ruotolo, B.T.; Hausinger, R.P. Analysis of a Soluble (UreD:UreF:UreG)2 Accessory Protein Complex and Its Interactions with Klebsiella aerogenes Urease by Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2013, 24, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Moncrief, M.B.C.; Hausinger, R.P. Characterization of UreG, Identification of a UreD-UreF-UreG Complex, and Evidence Suggesting That a Nucleotide-Binding Site in UreG Is Required for in vivo Metallocenter Assembly of Klebsiella aerogenes Urease. J. Bacteriol. 1997, 179, 4081–4086. [Google Scholar] [CrossRef]
- Boer, J.L.; Quiroz-Valenzuela, S.; Anderson, K.L.; Hausinger, R.P. Mutagenesis of Klebsiella aerogenes UreG to Probe Nickel Binding and Interactions with Other Urease-Related Proteins. Biochemistry 2010, 49, 5859–5869. [Google Scholar] [CrossRef]
- Rain, J.C.; Selig, L.; De Reuse, H.; Battaglia, V.; Reverdy, C.; Simon, S.; Lenzen, G.; Petel, F.; Wojcik, J.; Schächter, V.; et al. The Protein-Protein Interaction Map of Helicobacter pylori. Nature 2001, 409, 211–215. [Google Scholar] [CrossRef]
- Fong, Y.H.; Wong, H.C.; Chuck, C.P.; Chen, Y.W.; Sun, H.; Wong, K.-B. Assembly of Preactivation Complex for Urease Maturation in Helicobacter pylori: Crystal Structure of UreF-UreH Protein Complex. J. Biol. Chem. 2011, 286, 43241–43249. [Google Scholar] [CrossRef]
- Lam, R.; Romanov, V.; Johns, K.; Battaile, K.P.; Wu-Brown, J.; Guthrie, J.L.; Hausinger, R.P.; Pai, E.F.; Chirgadze, N.Y. Crystal Structure of a Truncated Urease Accessory Protein UreF from Helicobacter pylori. Proteins 2010, 78, 2839–2848. [Google Scholar] [CrossRef]
- Boer, J.L.; Hausinger, R.P. Klebsiella aerogenes UreF: Identification of the UreG Binding Site and Role in Enhancing the Fidelity of Urease Activation. Biochemistry 2012, 51, 2298–2308. [Google Scholar] [CrossRef]
- Yuen, M.H.; Fong, Y.H.; Nim, Y.S.; Lau, P.H.; Wong, K.-B. Structural Insights into how GTP-Dependent Conformational Changes in a Metallochaperone UreG Facilitate Urease Maturation. Proc. Natl. Acad. Sci. USA 2017, 114, E10890–E10898. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, H.; Lai, T.-P.; Sun, H. UreE-UreG Complex Facilitates Nickel Transfer and Preactivates GTPase of UreG in Helicobacter pylori. J. Biol. Chem. 2015, 290, 12474–12485. [Google Scholar] [CrossRef] [PubMed]
- Leipe, D.D.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. Classification and Evolution of P-Loop GTPases and Related ATPases. J. Mol. Biol. 2002, 317, 41–72. [Google Scholar] [CrossRef] [PubMed]
- Zambelli, B.; Turano, P.; Musiani, F.; Neyroz, P.; Ciurli, S. Zn2+-Linked Dimerization of UreG from Helicobacter pylori, a Chaperone Involved in Nickel Trafficking and Urease Activation. Proteins Struct. Funct. Bioinform. 2009, 74, 222–239. [Google Scholar] [CrossRef] [PubMed]
- Zambelli, B.; Stola, M.; Musiani, F.; De Vriendt, K.; Samyn, B.; Devreese, B.; Van Beeumen, J.; Turano, P.; Dikiy, A.; Bryant, D.A.; et al. UreG, a Chaperone in the Urease Process, Is an Intrinsically Unstructured Protein Binding a Single Zn2+ Ion. J. Biol. Chem. 2005, 280, 4684–4695. [Google Scholar] [CrossRef] [PubMed]
- Real-Guerra, R.; Staniscuaski, F.; Zambelli, B.; Musiani, F.; Ciurli, S.; Carlini, C.R. Biochemical and Structural Studies on Native and Recombinant Glycine max UreG: A Detailed Characterization of a Plant Urease Accessory Protein. Plant Mol. Biol. 2012, 78, 461–475. [Google Scholar] [CrossRef] [PubMed]
- Miraula, M.; Ciurli, S.; Zambelli, B. Intrinsic Disorder and Metal Binding in UreG Proteins from Archae Hyperthermophiles: GTPase Enzymes Involved in the Activation of Ni(II) Dependent Urease. J. Biol. Inorg. Chem. 2015, 20, 739–755. [Google Scholar] [CrossRef] [PubMed]
- Martin-Diaconescu, V.; Bellucci, M.; Musiani, F.; Ciurli, S.; Maroney, M.J. Unraveling the Helicobacter pylori UreG Zinc Binding Site Using X-Ray Absorption Spectroscopy (XAS) and Structural Modeling. J. Biol. Inorg. Chem. 2012, 17, 353–361. [Google Scholar] [CrossRef]
- Xia, W.; Li, H.; Yang, X.; Wong, K.B.; Sun, H. Metallo-GTPase HypB from Helicobacter pylori and Its Interaction with Nickel Chaperone Protein HypA. J. Biol. Chem. 2012, 287, 6753–6763. [Google Scholar] [CrossRef] [PubMed]
- Lacasse, M.J.; Douglas, C.D.; Zamble, D.B. Mechanism of Selective Nickel Transfer from HypB to HypA, Escherichia coli [NiFe]-Hydrogenase Accessory Proteins. Biochemistry 2016, 55, 6821–6831. [Google Scholar] [CrossRef] [PubMed]
- Gasper, R.; Scrima, A.; Wittinghofer, A. Structural Insights into HypB, a GTP-Binding Protein That Regulates Metal Binding. J. Biol. Chem. 2006, 281, 27492–27502. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.; Ngu, T.T.; Kaluarachchi, H.; Zamble, D.B. Relationship between the GTPase, Metal-Binding, and Dimerization Activities of E. Coli HypB. J. Inorg. Biochem. 2011, 16, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.H.; Lee, K.M.; Wong, K.B. Interaction between Hydrogenase Maturation Factors HypA and HypB Is Required for [NiFe]-Hydrogenase Maturation. PLoS ONE 2012, 7, e32592. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.H.; Li, T.; Wong, C.O.; Wong, K.B. Structural Basis for GTP-Dependent Dimerization of Hydrogenase Maturation Factor HypB. PLoS ONE 2012, 7, e30547. [Google Scholar] [CrossRef] [PubMed]
- Remaut, H.; Safarov, N.; Ciurli, S.; Van Beeumen, J. Structural Basis for Ni2+ Transport and Assembly of the Urease Active Site by the Metallochaperone UreE from Bacillus pasteurii. J. Biol. Chem. 2001, 276, 49365–49370. [Google Scholar] [CrossRef]
- Song, H.K.; Mulrooney, S.B.; Huber, R.; Hausinger, R.P. Crystal Structure of Klebsiella aerogenes UreE, a Nickel-Binding Metallochaperone for Urease Activation. J. Biol. Chem. 2001, 276, 49359–49364. [Google Scholar] [CrossRef]
- Banaszak, K.; Martin-Diaconescu, V.; Bellucci, M.; Zambelli, B.; Rypniewski, W.; Maroney, M.J.; Ciurli, S. Crystallographic and X-Ray Absorption Spectroscopic Characterization of Helicobacter pylori UreE Bound to Ni2+ and Zn2+ Reveals a Role for the Disordered C-Terminal Arm in Metal Trafficking. Biochem. J. 2012, 441, 1017–1026. [Google Scholar] [CrossRef]
- Shi, R.; Munger, C.; Asinas, A.; Benoit, S.L.; Miller, E.; Matte, A.; Maier, R.J.; Cygler, M. Crystal Structures of apo and Metal-Bound Forms of the UreE Protein from Helicobacter pylori: Role of Multiple Metal Binding Sites. Biochemistry 2010, 49, 7080–7088. [Google Scholar] [CrossRef]
- Stola, M.; Musiani, F.; Mangani, S.; Turano, P.; Safarov, N.; Zambelli, B.; Ciurli, S. The Nickel Site of Bacillus pasteurii UreE, a Urease Metallo-Chaperone, as Revealed by Metal-Binding Studies and X-Ray Absorption Spectroscopy. Biochemistry 2006, 45, 6495–6509. [Google Scholar] [CrossRef]
- Lee, M.H.; Pankratz, H.S.; Wang, S.; Scott, R.A.; Finnegan, M.G.; Johnson, M.K.; Ippolito, J.A.; Christianson, D.W.; Hausinger, R.P. Purification and Characterization of Klebsiella aerogenes UreE Protein: A Nickel-binding Protein That Functions in Urease Metallocenter Assembly. Protein Sci. 1993, 2, 1042–1052. [Google Scholar] [CrossRef]
- Bellucci, M.; Zambelli, B.; Musiani, F.; Turano, P.; Ciurli, S. Helicobacter pylori UreE, a Urease Accessory Protein: Specific Ni2+- and Zn2+-Binding Properties and Interaction with Its Cognate UreG. Biochem. J. 2009, 422, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Grossoehme, N.E.; Mulrooney, S.B.; Hausinger, R.P.; Wilcox, D.E. Thermodynamics of Ni2+, Cu2+, and Zn2+ Binding to the Urease Metallochaperone. Biochemistry 2007, 46, 10506–10516. [Google Scholar] [CrossRef] [PubMed]
- Won, H.; Lee, Y.; Kim, J.; Shin, I.S.; Lee, M.H.; Lee, B. Structural Characterization of the Nickel-Binding Properties of Bacillus pasteurii Urease Accessory Protein (Ure)E in Solution. J. Biol. Chem. 2004, 279, 17466–17472. [Google Scholar] [CrossRef] [PubMed]
- Ciurli, S.; Safarov, N.; Miletti, S.; Dikiy, A.; Christensen, S.K.; Kornetzky, K.; Bryant, D.A.; Vandenberghe, I.; Devreese, B.; Samyn, B.; et al. Molecular Characterization of Bacillus pasteurii UreE, a Metal-Binding Chaperone for the Assembly of the Urease Active Site. J. Biol. Inorg. Chem. 2002, 7, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Colpas, G.J.; Hausinger, R.P. In vivo and in citro Kinetics of Metal Transfer by the Klebsiella aerogenes Urease Nickel Metallochaperone, UreE. J. Biol. Chem. 2000, 275, 10731–10737. [Google Scholar] [CrossRef] [PubMed]
- Colpas, G.J.; Brayman, T.G.; Ming, L.J.; Hausinger, R.P. Identification of Metal-Binding Residues in the Klebsiella aerogenes Urease Nickel Metallochaperone, UreE. Biochemistry 1999, 38, 4078–4088. [Google Scholar] [CrossRef] [PubMed]
- Brayman, T.G.; Hausinger, R.P. Purification, Characterization, and Functional Analysis of a Truncated Klebsiella aerogenes UreE Urease Accessory Protein Lacking the Histidine-Rich Carboxyl Terminus. J. Bacteriol. 1996, 178, 5410–5416. [Google Scholar] [CrossRef] [PubMed]
- Mulrooney, S.B.; Ward, S.K.; Hausinger, R.P. Purification and Properties of the Klebsiella aerogenes UreE Metal-Binding Domain, a Functional Metallochaperone of Urease. J. Bacteriol. 2005, 187, 3581–3585. [Google Scholar] [CrossRef]
- Musiani, F.; Zambelli, B.; Stola, M.; Ciurli, S. Nickel Trafficking: Insights into the Fold and Function of UreE, a Urease Metallochaperone. J. Inorg. Biochem. 2004, 98, 803–813. [Google Scholar] [CrossRef]
- Merloni, A.; Dobrovolska, O.; Zambelli, B.; Agostini, F.; Bazzani, M.; Musiani, F.; Ciurli, S. Molecular Landscape of the Interaction between the Urease Accessory Proteins UreE and UreG. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 1662–1674. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, M.A.; Wang, B.; Feig, M.; Hausinger, R.P. Mutational and Computational Evidence That a Nickel-Transfer Tunnel in UreD Is Used for Activation of Klebsiella aerogenes Urease. Biochemistry 2015, 54, 6392–6401. [Google Scholar] [CrossRef] [PubMed]
- Soriano, A.; Colpas, G.J.; Hausinger, R.P. UreE Stimulation of GTP-Dependent Urease Activation in the UreD-UreF-UreG-Urease Apoprotein Complex. Biochemistry 2000, 39, 12435–12440. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Kuchar, J.; Hausinger, R.P. Chemical Cross-Linking and Mass Spectrometric Identification of Sites of Interaction for UreD, UreF, and Urease. J. Biol. Chem. 2004, 279, 15305–15313. [Google Scholar] [CrossRef]
- Musiani, F.; Gioia, D.; Masetti, M.; Falchi, F.; Cavalli, A.; Recanatini, M.; Ciurli, S. Protein Tunnels: The Case of Urease Accessory Proteins. J. Chem. Theory Comput. 2017, 13, 2322–2331. [Google Scholar] [CrossRef]
- Zambelli, B.; Berardi, A.; Martin-Diaconescu, V.; Mazzei, L.; Musiani, F.; Maroney, M.J.; Ciurli, S. Nickel Binding Properties of Helicobacter pylori UreF, an Accessory Protein in the Nickel-Based Activation of Urease. J. Biol. Inorg. Chem. 2014, 19, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Baykov, A.A.; Evtushenko, O.A.; Avaeva, S.M. A Malachite Green Procedure for Orthophosphate Determination and Its Use in Alkaline Phosphatase-Based Enzyme Immunoassay. Anal. Biochem. 1988, 171, 266–270. [Google Scholar] [CrossRef]
- Maier, T.; Lottspeich, F.; Böck, A. GTP Hydrolysis by HypB Is Essential for Nickel Insertion into Hydrogenases of Escherichia coli. Eur. J. Biochem. 1995, 230, 133–138. [Google Scholar] [CrossRef]
- Olson, J.W.; Fu, C.; Maier, R.J. The HypB Protein from Bradyrhizobium japonicum can Store Nickel and Is Required for the Nickel-dependent Transcriptional Regulation of Hydrogenase. Mol. Microbiol. 1997, 24, 119–128. [Google Scholar] [CrossRef]
- Olson, J.W.; Mehta, N.S.; Maier, R.J. Requirement of Nickel Metabolism Proteins HypA and HypB for Full Activity of Both Hydrogenase and Urease in Helicobacter pylori. Mol. Microbiol. 2001, 39, 176–182. [Google Scholar] [CrossRef]
- Hube, M.; Blokesch, M.; Böck, A. Network of Hydrogenase Maturation in Escherichia coli: Role of Accessory Proteins HypA and HybF. J. Bacteriol. 2002, 184, 3879–3885. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, D.; Gutekunst, K.; Klissenbauer, M.; Schulz-Friedrich, R.; Appel, J. Mutagenesis of Hydrogenase Accessory Genes of Synechocystis sp. PCC 6803: Additional Homologues of hypA and hypB Are Not Active in Hydrogenase Maturation. FEBS J. 2006, 273, 4516–4527. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Jacobi, A.; Sauter, M.; Böck, A. The Product of the HypB Gene, Which Is Required for Nickel Incorporation into Hydrogenases, Is a Novel Guanine Nucleotide-Binding Protein. J. Bacteriol. 1993, 175, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, A.; Rossmann, R.; Böck, A. The hyp Operon Gene Products Are Required for the Maturation of Catalytically Active Hydrogenase Isoenzymes in Escherichia coli. Arch. Microbiol. 1992, 158, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Lutz, S.; Jacobi, A.; Schlensog, V.; Böhm, R.; Sawers, G.; Böck, A. Molecular Characterization of an Operon (hyp) Necessary for the Activity of the Three Hydrogenase Isoenzymes in Escherichia coli. Mol. Microbiol. 1991, 5, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Tomb, J.F.; White, O.; Kerlavage, A.R.; Clayton, R.A.; Sutton, G.G.; Fleischmann, R.D.; Ketchum, K.A.; Klenk, H.P.; Gill, S.; Brian, A.D. The Complete Genome Sequence of the Gastric Pathogen Helicobacter pylori. Nature 1997, 388, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Spronk, C.A.E.M.; Żerko, S.; Górka, M.; Koźmiński, W.; Bardiaux, B.; Zambelli, B.; Musiani, F.; Piccioli, M.; Basak, P.; Blum, F.C.; et al. Structure and Dynamics of Helicobacter pylori Nickel-chaperone HypA: An Integrated Approach Using NMR Spectroscopy, Functional Assays and Computational Tools. JBIC J. Biol. Inorg. Chem. 2018, 23, 1309–1330. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Li, H.; Sze, K.H.; Sun, H. Structure of a Nickel Chaperone, HypA, from Helicobacter pylori Reveals Two Distinct Metal Binding Sites. J. Am. Chem. Soc. 2009, 131, 10031–10040. [Google Scholar] [CrossRef]
- Mehta, N.; Olson, J.W.; Maier, R.J. Characterization of Helicobacter pylori Nickel Metabolism Accessory Proteins Needed for Maturation of Both Urease and Hydrogenase. J. Bacteriol. 2003, 185, 726–734. [Google Scholar] [CrossRef]
- Herbst, R.W.; Perovic, I.; Martin-diaconescu, V.; Brien, K.O.; Chivers, P.T.; Pochapsky, S.S.; Pochapsky, T.C.; Maroney, M.J. Communication between the Zinc and Nickel Sites in Dimeric HypA: Metal Recognition and pH Sensing. J. Am. Chem. Soc. 2010, 132, 10338–10351. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.Q.; Johnson, R.C.; Merrell, D.S.; Maroney, M.J. Nickel Ligation of the N-Terminal Amine of HypA Is Required for Urease Maturation in Helicobacter pylori. Biochemistry 2017, 56, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Atanassova, A.; Zamble, D.B. Escherichia coli HypA Is a Zinc Metalloprotein with a Weak Affinity for Nickel. J. Bacteriol. 2005, 187, 4689–4697. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Arai, T.; Matsumi, R.; Atomi, H.; Imanaka, T.; Miki, K. Crystal Structure of HypA, a Nickel-Binding Metallochaperone for [NiFe] Hydrogenase Maturation. J. Mol. Biol. 2009, 394, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.C.; Herbst, R.W.; Iwig, J.S.; Chivers, P.T.; Maroney, M.J. A Dynamic Zn Site in Helicobacter pylori HypA: A Potential Mechanism for Metal-Specific Protein Activity. J. Am. Chem. Soc. 2007, 129, 16–17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.C.; Hu, H.Q.; Merrell, D.S.; Maroney, M.J. Dynamic HypA Zinc Site Is Essential for Acid Viability and Proper Urease Maturation in Helicobacter pylori. Metallomics 2015, 7, 674–682. [Google Scholar] [CrossRef]
- Stingl, K.; Schauer, K.; Ecobichon, C.; Labigne, A.; Lenormand, P.; Rousselle, J.-C.; Namane, A.; de Reuse, H. In vivo Interactome of Helicobacter pylori Urease Revealed by Tandem Affinity Purification. Mol. Cell. Proteom. 2008, 7, 2429–2441. [Google Scholar] [CrossRef]
- Hu, H.Q.; Huang, H.T.; Maroney, M.J. The Helicobacter pylori HypA·UreE2 Complex Contains a Novel High-Affinity Ni(II)-Binding Site. Biochemistry 2018, 57, 2932–2942. [Google Scholar] [CrossRef]
- Yang, X.; Li, H.; Cheng, T.; Xia, W.; Lai, Y.-T.; Sun, H. Nickel Translocation between Metallochaperones HypA and UreE in Helicobacter pylori. Metallomics 2014, 6, 40–42. [Google Scholar] [CrossRef]
- Benoit, S.L.; McMurry, J.L.; Hill, S.A.; Maier, R.J. Helicobacter pylori Hydrogenase Accessory Protein HypA and Urease Accessory Protein UreG Compete with Each Other for UreE Recognition. Biochim. Biophys. Acta-Gen. Subj. 2012, 1820, 1519–1525. [Google Scholar] [CrossRef]
- Douglas, C.D.; Ngu, T.T.; Kaluarachchi, H.; Zamble, D.B. Metal Transfer within the Escherichia coli HypB−HypA Complex of Hydrogenase Accessory Proteins. Biochemistry 2013, 52, 6030–6039. [Google Scholar] [CrossRef]
- Kaluarachchi, H.; Zhang, J.W.; Zamble, D.B. Escherichia coli SlyD, More Than a Ni(II) Reservoir. Biochemistry 2011, 50, 10761–10763. [Google Scholar] [CrossRef]
- Cheng, T.; Li, H.; Yang, X.; Xia, W.; Sun, H. Interaction of SlyD with HypB of Helicobacter pylori Facilitates Nickel Trafficking. Metallomics 2013, 5, 804–807. [Google Scholar] [CrossRef]
- Zhang, J.W.; Butland, G.; Greenblatt, J.F.; Emili, A.; Zamble, D.B. A Role for SlyD in the Escherichia coli Hydrogenase Biosynthetic Pathway. J. Biol. Chem. 2005, 280, 4360–4366. [Google Scholar] [CrossRef]
- Leach, M.R.; Jie, W.Z.; Zamble, D.B. The Role of Complex Formation between the Escherichia coli Hydrogenase Accessory Factors HypB and SlyD. J. Biol. Chem. 2007, 282, 16177–16186. [Google Scholar] [CrossRef]
- Lacasse, M.J.; Zamble, D.B. [NiFe]-Hydrogenase Maturation. Biochemistry 2016, 55, 1689–1701. [Google Scholar] [CrossRef]
- Zeer-wanklyn, C.J.; Zamble, D.B. Microbial Nickel: Cellular Uptake and Delivery to Enzyme Centers. Curr. Opin. Chem. Biol. 2017, 37, 80–88. [Google Scholar] [CrossRef]
- Witte, C.-P. Urea Metabolism in Plants. Plant Sci. 2011, 180, 431–438. [Google Scholar] [CrossRef]
- Polacco, J.C.; Mazzafera, P.; Tezotto, T. Opinion—Nickel and Urease in Plants: Still Many Knowledge Gaps. Plant Sci. 2013, 199–200, 79–90. [Google Scholar] [CrossRef]
- Cao, F.-Q.; Werner, A.K.; Dahncke, K.; Romeis, T.; Liu, L.-H.; Witte, C.-P. Identification and Characterization of Proteins Involved in Rice Urea and Arginine Catabolism. Plant Physiol. 2010, 154, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Myrach, T.; Zhu, A.; Witte, C.-P. The Assembly of the Plant Urease Activation Complex and the Essential Role of the Urease Accessory Protein G (UreG) in Delivery of Nickel to Urease. J. Biol. Chem. 2017, 292, 14556–14565. [Google Scholar] [CrossRef]
- Witte, C.-P.; Rosso, M.G.; Romeis, T. Identification of Three Urease Accessory Proteins that are Required for Urease Activation in Arabidopsis. Plant Physiol. 2005, 139, 1155–1162. [Google Scholar] [CrossRef]
- Freyermuth, S.K.; Bacanamwo, M.; Polacco, J.C. The Soybean Eu3 Gene Encodes an Ni-Binding Protein Necessary for Urease Activity. Plant J. 2000, 21, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, J.C. The Emerging Role of Urease as a General Microbial Virulence Factor. PLoS Pathog. 2014, 10, e1004062. [Google Scholar] [CrossRef] [PubMed]
- Safavi, M.; Sabourian, R.; Foroumadi, A. Treatment of Helicobacter pylori Infection: Current and Future Insights. World J. Clin. Cases 2016, 4, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Koohi-moghadam, M.; Wang, R.; Chang, Y.; Woo, P.C.Y.; Wang, J.; Li, H.; Sun, H. Metallochaperone UreG Serves as a New Target for Design of Urease Inhibitor: A Novel Strategy for Development of Antimicrobials. PLoS Biol. 2018, 16, e2003887. [Google Scholar] [CrossRef] [PubMed]
- Eschweiler, J.D.; Farrugia, M.A.; Dixit, S.M.; Hausinger, R.P.; Ruotolo, B.T. A Structural Model of the Urease Activation Complex Derived from Ion Mobility-Mass Spectrometry and Integrative Modeling. Structure 2018, 26, 599–606.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ligabue-Braun, R.; Real-Guerra, R.; Carlini, C.R.; Verli, H. Evidence-Based Docking of the Urease Activation Complex. J. Biomol. Struct. Dyn. 2013, 31, 854–861. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nim, Y.S.; Wong, K.-B. The Maturation Pathway of Nickel Urease. Inorganics 2019, 7, 85. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics7070085
Nim YS, Wong K-B. The Maturation Pathway of Nickel Urease. Inorganics. 2019; 7(7):85. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics7070085
Chicago/Turabian StyleNim, Yap Shing, and Kam-Bo Wong. 2019. "The Maturation Pathway of Nickel Urease" Inorganics 7, no. 7: 85. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics7070085



