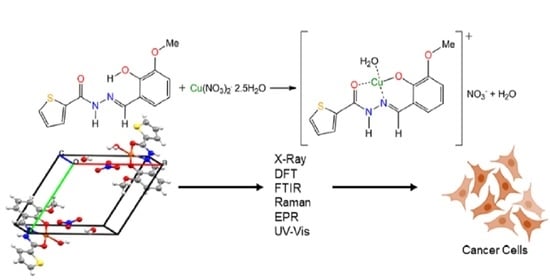
Synthesis, Crystal Structure, Spectroscopic Characterization, DFT Calculations and Cytotoxicity Assays of a New Cu(II) Complex with an Acylhydrazone Ligand Derived from Thiophene
Abstract
:1. Introduction
2. Results and Discussion
2.1. Syntheses
2.2. Crystal Structure
2.3. Vibrational Spectroscopy
2.4. EPR Spectroscopy
2.5. Electronic Spectroscopy
2.6. Cell Viability Study
2.7. Antioxidant Activity
3. Materials and Methods
3.1. Synthesis
3.2. X-ray Diffraction Data
3.3. Spectroscopy
3.4. Computational Methods
3.5. Cell Viability Study: 3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide Assay
Cell Line and Growth Conditions
3.6. Antioxidant Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linder, M.C.; Hazegh-Azam, M. Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 1996, 63, 797S–811S. [Google Scholar] [PubMed]
- Crisponi, G.; Nurchi, V.M.; Fanni, D.; Gerosa, C.; Nemolato, S.; Faa, G. Copper-related diseases: From chemistry to molecular pathology. Coord. Chem. Rev. 2010, 254, 876–889. [Google Scholar] [CrossRef]
- Peña, M.M.O.; Lee, J.; Thiele, D.J. A Delicate Balance: Homeostatic Control of Copper Uptake and Distribution. J. Nutr. 1999, 129, 1251–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, I.; Mahmood, L.M.A.; Mehdar, Y.T.H.; Aboul-Enein, H.Y.; Said, M.A. Synthesis, characterization, simulation, DNA binding and anticancer activities of Co(II), Cu(II), Ni(II) and Zn(II) complexes of a Schiff base containing o-hydroxyl group nitrogen ligand. Inorg. Chem. Commun. 2020, 118, 108004. [Google Scholar] [CrossRef]
- Gündüzalp, A.B.; Özsen, I.; Alyar, H.; Alyar, S.; Özbek, N. Biologically active Schiff bases containing thiophene/furan ring and their copper(II) complexes: Synthesis, spectral, nonlinear optical and density functional studies. J. Mol. Struct. 2016, 1120, 259–266. [Google Scholar] [CrossRef]
- Puszko, A.; Krojcer, A.; Pełczynska, M.; Wietrzyk, J.; Cieślak-Golonka, M.; Jezierska, J.; Adach, A.; Kubiak, M. Mononuclear copper(II) nitrato complexes with methyl-substituted 4-nitropyridine N-oxide. Physicochemical and cytotoxic characteristics. J. Inorg. Biochem. 2010, 104, 153–160. [Google Scholar] [CrossRef]
- Syamal, A.; Maurya, M.R.; Dooms, E.; McLean, J.A. Synthesis and Characterization of Nickel (II), Cobalt (II), Copper(II), Manganese (II), Zinc (II), Zirconium (IV), Oxomolybdenum (V), and Dioxouranium (VI) Complexes of the Schiff Base Derived from Salycylaldehyde and Thiophene-2-Carboxylic Acid Hydrazide. Synth. React. Inorg. Met. Chem. 1986, 16, 39–60. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in copper complexes as anticancer agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef]
- Weder, J.E.; Dillon, C.T.; Hambley, T.W.; Kennedy, B.J.; Lay, P.A.; Biffin, J.R.; Regtop, H.L.; Davies, N.M. Copper Complexes of Non-Steroidal Anti-Inflammatory Drugs: An Opportunity Yet to Be Realized. Coord. Chem. Rev. 2002, 223, 95–126. [Google Scholar] [CrossRef]
- Rodríguez-Argüelles, M.C.; Mosquera-Vázquez, S.; Tourón-Touceda, P.; Sanmartín-Matalobos, J.; García-Deibe, A.M.; Belicchi-Ferrari, M.; Pelosi, G.; Pelizzi, C.; Zani, F. Complexes of 2-thiophenecarbonyl and isonicotinoyl hydrazones of 3-(N-methyl)isatin. A study of their antimicrobial activity. J. Inorg. Biochem. 2007, 101, 138–147. [Google Scholar] [CrossRef]
- Anitha, C.; Sheela, C.D.; Tharmaraj, P.; Johnson Raja, S. Synthesis and characterization of VO(II), Co(II), Ni(II), Cu(II) and Zn(II) complexes of chromone based azo-linked Schiff base ligand. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 98, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Manzur, C.; Novoa, N.; Celedón, S.; Carrillo, D.; Hamon, J.R. Multidentate unsymmetrically-substituted Schiff bases and their metal complexes: Synthesis, functional materials properties, and applications to catalysis. Coord. Chem. Rev. 2018, 357, 144–172. [Google Scholar] [CrossRef]
- Da Silva, C.M.; Da Silva, D.L.; Modolo, L.V.; Alves, R.B.; De Resende, M.A.; Martins, C.V.B.; De Fátima, Â. Schiff bases: A short review of their antimicrobial activities. J. Adv. Res. 2011, 2, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Kajal, A.; Bala, S.; Kamboj, S.; Sharma, N.; Saini, V. Schiff Bases: A Versatile Pharmacophore. J. Catal. 2013, 2013, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Desai, S.Β.; Desai, P.B.; Desai, K.R. Syntheses of some Schiff bases, thiazolididones and azetidinones derived from 2,6-diaminobenzo[1,2-d:4,5-d’] bisthiazole and their anticancer activities. Heterocycl. Commun. 2001, 7. [Google Scholar] [CrossRef]
- Jarrahpour, A.; Motamedifar, M.; Pakshir, K.; Hadi, N.; Zarei, M. Synthesis of Novel Azo Schiff Bases and Their Antibacterial and Antifungal Activities. Molecules 2004, 9, 815–824. [Google Scholar] [CrossRef] [Green Version]
- Hearn, M.J.; Cynamon, M.H.; Chen, M.F.; Coppins, R.; Davis, J.; Joo-On Kang, H.; Noble, A.; Tu-Sekine, B.; Terrot, M.S.; Trombino, D.; et al. Preparation and antitubercular activities in vitro and in vivo of novel Schiff bases of isoniazid. Eur. J. Med. Chem. 2009, 44, 4169–4178. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.J.; Qin, X.; Liu, K.; Zhu, D.D.; Wang, X.M.; Zhu, H.L. Synthesis, antibacterial activities and molecular docking studies of Schiff bases derived from N-(2/4-benzaldehyde-amino) phenyl-N′-phenyl-thiourea. Bioorganic Med. Chem. 2011, 19, 5708–5715. [Google Scholar] [CrossRef]
- Hameed, A.; al-Rashida, M.; Uroos, M.; Abid Ali, S.; Khan, K.M. Schiff bases in medicinal chemistry: A patent review (2010–2015). Expert Opin. Ther. Pat. 2017, 27, 63–79. [Google Scholar] [CrossRef]
- Przybylski, P.; Huczynski, A.; Pyta, K.; Brzezinski, B.; Bartl, F. Biological Properties of Schiff Bases and Azo Derivatives of Phenols. Curr. Org. Chem. 2009, 13, 124–148. [Google Scholar] [CrossRef]
- Nogueira, L.; Cardoso, D.F.; Cristina, T.; Nogueira, M.; Kaiser, C.R.; Wardell, J.L.; Maria, S.; Veloso, S.; Vinicius, M.; Souza, N. De Synthesis and anti-tubercular activity of Thienyl and Furanyl derivatives. Mediterr. J. Chem. 2016, 5, 356–366. [Google Scholar]
- Mohareb, R.M.; Fleita, D.H.; Sakka, O.K. Novel synthesis of hydrazide-hydrazone derivatives and their utilization in the synthesis of coumarin, pyridine, thiazole and thiophene derivatives with antitumor activity. Molecules 2011, 16, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Baró, A.C.; Pis-Diez, R.; Parajón-Costa, B.S.; Rey, N.A. Spectroscopic and theoretical study of the o-vanillin hydrazone of the mycobactericidal drug isoniazid. J. Mol. Struct. 2012, 1007, 95–101. [Google Scholar] [CrossRef]
- Rodríguez, M.R.; Del Plá, J.; Piro, O.E.; Echeverría, G.A.; Espino, G.; Pis-Diez, R.; Parajón-Costa, B.S.; González-Baró, A.C. Structure, tautomerism, spectroscopic and DFT study of o-vanillin derived Schiff bases containing thiophene ring. J. Mol. Struct. 2018, 1165, 381–390. [Google Scholar] [CrossRef]
- Mohamed, G.G.; Sharaby, C.M. Metal complexes of Schiff base derived from sulphametrole and o-vanilin. Synthesis, spectral, thermal characterization and biological activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2007, 66, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-Y.; Xian, H.-D.; Liu, J.-F.; Zhao, G.-L. Synthesis, Characterization, Crystal Structure and Antibacterial Activities of Transition Metal(II) Complexes of the Schiff Base 2-[(4-Methylphenylimino)methyl]-6-methoxyphenol. Molecules 2009, 14, 1747–1754. [Google Scholar] [CrossRef] [Green Version]
- Mishra, R.; Jha, K.K.; Kumar, S.; Tomer, I. Synthesis, properties and biological activity of thiophene: A review. Pharma Chem. 2011, 3, 38–54. [Google Scholar]
- Malik, M.A.; Dar, O.A.; Gull, P.; Wani, M.Y.; Hashmi, A.A. Heterocyclic Schiff base transition metal complexes in antimicrobial and anticancer chemotherapy. Medchemcomm 2018, 9, 409–436. [Google Scholar] [CrossRef]
- Forsch, R.A.; Wright, J.E.; Rosowsky, A. Synthesis and In Vitro Antitumor Activity of Thiophene Analogues of 5-Chloro-5,8-dideazafolic Acid and 2-methyl-2-desamino-5-chloro-5,8-dideazafolic acid. Bioorg. Med. Chem. 2002, 10, 2067–2076. [Google Scholar] [CrossRef]
- Cardoso, L.; Nogueira, T.; Rodrigues, F.; Oliveira, A.; Dos Santos Luciano, C.; Pessoa, C.; De Souza, M. CHEMISTRY N-acylhydrazones containing thiophene nucleus: A new anticancer class. Med. Chem. Res. 2017, 1–4. [Google Scholar] [CrossRef]
- Molvi, K.I.; Vasu, K.K.; Yerande, S.G.; Sudarsanam, V.; Haque, N. Syntheses of new tetrasubstituted thiophenes as novel anti-inflammatory agents. Eur. J. Med. Chem. 2007, 42, 1049–1058. [Google Scholar] [CrossRef]
- Jha, K.; Kumar, S.; Tomer, I.; Mishra, R. Thiophene: The molecule of diverse medicinal importance. J. Pharm. Res 2012, 5, 560–566. [Google Scholar]
- Duffy, J.L.; Kirk, B.A.; Konteatis, Z.; Campbell, E.L.; Liang, R.; Brady, E.J.; Candelore, M.R.; Ding, V.D.H.; Jiang, G.; Liu, F.; et al. Discovery and investigation of a novel class of thiophene-derived antagonists of the human glucagon receptor. Bioorg. Med. Chem. Lett. 2005, 15, 1401–1405. [Google Scholar] [CrossRef]
- Wardakhan, W.W.; Abdel-Salam, O.M.E.; Elmegeed, G.A. Screening for antidepressant, sedative and analgesic activities of novel fused thiophene derivatives. Acta Pharm. 2008, 58, 1–14. [Google Scholar] [CrossRef] [Green Version]
- El-Salam, O.I.A.; Shalaby, A.M.; El-Sawy, A.A.; Elshihaby, S.; Abdulla, M. Synthesis of Some N-[(4-Substituted-1-Piperazinyl)-Oxo(Alkyl and Ethylcarbamoyl)]-3-(2-Thiophenyl)Acrylamides as Non-Steroidal Anti-Allergic and Anti-Inflammatory Agents. Open J. Synth. Theory Appl. 2013, 2, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Fadda, A.A.; Berghot, M.A.; Amer, F.A.; Badawy, D.S.; Bayoumy, N.M. Synthesis and antioxidant and antitumor activity of novel pyridine, chromene, thiophene and thiazole derivatives. Arch. Pharm. (Weinh.) 2012, 345, 378–385. [Google Scholar] [CrossRef]
- Farrugia, L.J. ORTEP-3 for Windows—A version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Crystallogr. 1997, 30, 565–566. [Google Scholar] [CrossRef]
- Addison, C.C.; Gatehouse, B.M. The infrared spectra of anhydrous transition-metal nitrates. J. Chem. Soc. 1960, 613. [Google Scholar] [CrossRef]
- Hathaway, B.J.; Billing, D.E. The electronic properties and stereochemistry of mono-nuclear complexes of the copper(II) ion. Coord. Chem. Rev. 1970, 5, 143–207. [Google Scholar] [CrossRef]
- Garribba, E.; Micera, G. The Determination of the Geometry of Cu(II) Complexes: An EPR Spectroscopy Experiment. J. Chem. Educ. 2006, 83, 1229. [Google Scholar] [CrossRef]
- González-Álvarez, M.; Alzuet, G.; Borrás, J.; Agudo, L.D.C.; García-Granda, S.; Montejo-Bernardo, J.M. Comparison of protective effects against reactive oxygen species of mononuclear and dinuclear Cu(II) Complexes with N-substituted benzothiazolesulfonamides. Inorg. Chem. 2005, 44, 9424–9433. [Google Scholar] [CrossRef] [PubMed]
- Billing, D.E.; Dudley, R.J.; Hathaway, B.J.; Tomlinson, A.A.G. Single-Crystal Electronic and Electron Spin Resonance Spectra of Di-chloroaquo-(2,9-dimethyl-1,10-phenanthroline)copper(II). J. Chem. Soc. 1971, 691–696. [Google Scholar] [CrossRef]
- Hathaway, B.J. The correlation of the electronic properties and stereochemistry of mononuclear {CuN 4–6 } chromophores. J. Chem. Soc. Dalt. Trans. 1972, 1196–1199. [Google Scholar] [CrossRef]
- Kortum, G. Reflectance Spectroscopy Principles, Methods, Applications; Springer: New York, NY, USA, 1969; ISBN 3642880711. [Google Scholar]
- Mei, Y.; Zhang, S.; Hu, C.; Zhang, J.; Yang, M. Synthesis, characterization, and crystal structures of mononuclear and dinuclear copper(II) complexes derived from similar tridentate Schiff bases. Inorg. Nano-Met. Chem. 2017, 47, 1270–1274. [Google Scholar] [CrossRef]
- Jayendran, M.; Begum, P.M.S.; Kurup, M.R.P. Structural, spectral and biological investigations on Cu(II) and Zn(II) complexes derived from NNO donor tridentate Schiff base: Crystal structure of a 1D Cu(II) coordination polymer. J. Mol. Struct. 2020, 1206, 127682. [Google Scholar] [CrossRef]
- Bhunia, A.; Vojtíšek, P.; Bertolasi, V.; Manna, S.C. Tridentate Schiff base coordinated trigonal bipyramidal/square pyramidal copper(II) complexes: Synthesis, crystal structure, DFT/TD-DFT calculation, catecholase activity and DNA binding. J. Mol. Struct. 2019, 1189, 94–101. [Google Scholar] [CrossRef]
- Benítez, J.; Guggeri, L.; Tomaz, I.; Pessoa, J.C.; Moreno, V.; Lorenzo, J.; Avilés, F.X.; Garat, B.; Gambino, D. A novel vanadyl complex with a polypyridyl DNA intercalator as ligand: A potential anti-protozoa and anti-tumor agent. J. Inorg. Biochem. 2009, 103, 1386–1394. [Google Scholar] [CrossRef]
- Leon, I.E.; Di Virgilio, A.L.; Porro, V.; Muglia, C.I.; Naso, L.G.; Williams, P.A.; Bollati-Fogolin, M.; Etcheverry, S.B. Antitumor properties of a vanadyl(IV) complex with the flavonoid chrysin [VO(chrysin)2EtOH]2 in a human osteosarcoma model: The role of oxidative stress and apoptosis. Dalton Trans. 2013, 42, 11868–11880. [Google Scholar] [CrossRef]
- Burgos-Lopez, Y.; Del Plá, J.; Balsa, L.M.; León, I.E.; Echeverría, G.A.; Piro, O.E.; García-Tojal, J.; Pis-Diez, R.; González-Baró, A.C.; Parajón-Costa, B.S. Synthesis, crystal structure and cytotoxicity assays of a copper(II) nitrate complex with a tridentate ONO acylhydrazone ligand. Spectroscopic and theoretical studies of the complex and its ligand. Inorg. Chim. Acta 2019, 487, 31–40. [Google Scholar] [CrossRef]
- Koňariková, K.; Perdikaris, G.A.; Gbelcová, H.; Andrezálová, L.; Švéda, M.; Ruml, T.; Laubertová, L.; Žitňanová, I. Effect of Schiff base Cu(II) complexes on signaling pathways in HT-29 cells. Mol. Med. Rep. 2016, 14, 4436–4444. [Google Scholar] [CrossRef] [Green Version]
- Hajrezaie, M.; Paydar, M.; Zorofchian Moghadamtousi, S.; Hassandarvish, P.; Gwaram, N.S.; Zahedifard, M.; Rouhollahi, E.; Karimian, H.; Looi, C.Y.; Ali, H.M.; et al. A Schiff Base-Derived Copper (II) Complex Is a Potent Inducer of Apoptosis in Colon Cancer Cells by Activating the Intrinsic Pathway. Sci. World J. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.D.; Su, H.; Zhao, J.; Zhao, B.X.; Zhang, S.L.; Miao, J.Y. A novel copper complex of salicylaldehyde pyrazole hydrazone induces apoptosis through up-regulating integrin β4 in H322 lung carcinoma cells. Eur. J. Med. Chem. 2010, 45, 1438–1446. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.R.; Balsa, L.M.; Del Plá, J.; García-Tojal, J.; Pis-Diez, R.; Parajón-Costa, B.S.; León, I.E.; González-Baró, A.C. Synthesis, characterization, DFT calculations and anticancer activity of a new oxidovanadium(IV) complex with a ligand derived from o-vanillin and thiophene. New J. Chem. 2019, 43, 11784–11794. [Google Scholar] [CrossRef]
- CrysAlisPro, Oxford Diffraction Ltd, version 1.171.33.48 (release 15-09-2009 CrysAlis171.NET); Oxford Diffraction Ltd: Abingdon-on-Thames, UK, 2009.
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Crystallogr. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- WinEPR SimFonia, Version 1.25; Bruker Analytische Messtechnik GmbH: Billerica, MA, USA, 1996.
- Yanai, T.; Tew, D.P.; Handy, N.C. A new hybrid exchange-correlation functional using the Coulomb-attenuating method (CAM-B3LYP). Chem. Phys. Lett. 2004, 393, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Neese, F. The ORCA program system. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 1999, 110, 6158–6170. [Google Scholar] [CrossRef]
- Klamt, A.; Schüürmann, G. COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc. Perkin Trans. 1993, 799–805. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant Activity Applying An Improved ABTS Radical Cation Decolorization Assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]







| Empirical formula | C13 H15 Cu N3 O8 S |
| Formula weight | 436.88 |
| Temperature | 293(2) K |
| Wavelength | 1.54184 Å |
| Crystal system | Triclinic |
| Space group | P-1 |
| Unit cell dimensions | |
| a | 9.3380(5) Å |
| b | 9.6015(5) Å |
| c | 11.3991(7) Å |
| α | 97.334(5)° |
| β | 104.595(5)° |
| ɣ | 116.924(5)° |
| Volume | 846.52(8) Å3 |
| Z, density (calculated) | 2, 1.714 Mg/m3 |
| Absorption coefficient | 3.442 mm−1 |
| F(000) | 446 |
| θ-range for data collection | 4.17 to 72.41° |
| Index ranges | −9 ≤ h ≤ 11, −11 ≤ k ≤ 9, −14 ≤ l ≤ 13 |
| Reflections collected | 7440 |
| Independent reflections | 3335 [R(int) = 0.023] |
| Observed reflections [I > 2σ(I)] | 2989 |
| Completeness to θ = 72.41° | 99.4% |
| Refinement method | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 3335/6/252 |
| Goodness-of-fit on F2 | 1.047 |
| Final R indices [I > 2σ(I)] | R1 = 0.0449, wR2 = 0.1264 |
| R indices (all data) | R1 = 0.0496, wR2 = 0.1323 |
| Largest diff. peak and hole | 0.641 and −0.555 e.Å−3 |
| Bond Distances (Å) | Bond Angles (°) | ||||
|---|---|---|---|---|---|
| Exp. | Calc. | Exp. | Calc. | ||
| Cu–N1 | 1.927 | 1.957 | N1–Cu–O3 | 81.6 | 79.4 |
| Cu–O1 | 1.885 | 1.915 | Cu–N1–N2 | 111.7 | 112.1 |
| Cu–O1W | 1.953 | 1.990 | N1–Cu–O1 | 92.8 | 91.1 |
| Cu–O3 | 1.978 | 2.094 | O1–Cu–O3 | 174.4 | 161.8 |
| Cu–O31 | 2.429 | 2.257 | O1W–Cu–N1 | 167.8 | 173.0 |
| O1–C1 | 1.311 | 1.285 | O1W–Cu–O1 | 92.4 | 94.5 |
| N1–C7 | 1.290 | 1.297 | N1–Cu–O31 | 98.0 | 92.3 |
| N1–N2 | 1.376 | 1.374 | O3–Cu–O1W | 93.1 | 94.0 |
| C8–N2 | 1.342 | 1.354 | O3–Cu–O31 | 96.6 | 89.2 |
| C8–O3 | 1.258 | 1.247 | O1–Cu–O31 | 84.2 | 106.9 |
| O31–N3 | 1.217 | 1.272 | O1W–Cu–O31 | 93.6 | 90.2 |
| C2–O2 | 1.365 | 1.353 | N3–O31–Cu | 136.4 | 127.0 |
| O2–C13 | 1.420 | 1.416 | |||
| N3–O32 | 1.212 | 1.275 | Dihedral Angles (°) | ||
| N3–O33 | 1.217 | 1.216 | Exp. | Calc. | |
| C6–C7 | 1.430 | 1.417 | O3–Cu–N1–N2 | −1.5 | −9.8 |
| N1–Cu–O1–C1 | 5.8 | −10.8 | |||
| O3–C8–C9–S | 4.9 | −10.2 | |||
| O3–C8–N2–N1 | −1.6 | 1.4 | |||
| O2–C2–C1–O1 | 0.9 | 0.0 | |||
| N2–C8–C9–S | −174.0 | 170.3 | |||
| Experimental in DMSO (ε, M−1·cm−1) | Experimental, Solid Sample | Calculated (O. S.) | Transition (a) | Assignment |
|---|---|---|---|---|
| 743 (43) | 715 ** | 843 (0.00023) | Hβ −9→ Lβ | d → d |
| 657 (35) | 637 ** | 670 (0.00040) | Hβ −15 → Lβ | d → d |
| 423 (sh) 404 (1.2 × 104) | 411 | 581 (0.00129) 573 (0.00159) 478 (0.10700) 438 (0.00123) | Hβ −16 → Lβ Hβ −17→ Lβ Hβ → Lβ Hβ −1 → Lβ | d → d d → d LMCT LMCT |
| 348 (sh) | 330 | 345 (0.30163) | Hα → Lα+1 | Intraligand |
| 335 (1.7 × 104) | 287 (0.48728) | Hβ −5 → Lβ | LMCT | |
| 320 (sh) | 264 (0.10541) | Hα −5 → Lα | Intraligand | |
| 258 * | 246 (0.02130) | Hα −1 → Lα+1 | Intraligand |
| Cell Lines | IC50 CuHL | SI CuHL | IC50 CDDP | SI CDDP |
|---|---|---|---|---|
| MG-63 | 2.0 ± 0.5 | 1.8 | 39 ± 1.8 | 0.3 |
| HT-29 | 7.4 ± 0.9 | 0.5 | 178.8 ± 4.9 | 0.06 |
| A549 | 3.1 ± 0.6 | 1.2 | 114.0 ± 2.3 | 0.1 |
| L929 | 3.6 ± 0.3 | 11.2 ± 1.6 |
| Compound | H2L | CuHL |
|---|---|---|
| TEAC | 1.33 | 0.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez, M.R.; Balsa, L.M.; Piro, O.E.; Etcheverría, G.A.; García-Tojal, J.; Pis-Diez, R.; León, I.E.; Parajón-Costa, B.P.; González-Baró, A.C. Synthesis, Crystal Structure, Spectroscopic Characterization, DFT Calculations and Cytotoxicity Assays of a New Cu(II) Complex with an Acylhydrazone Ligand Derived from Thiophene. Inorganics 2021, 9, 9. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics9020009
Rodríguez MR, Balsa LM, Piro OE, Etcheverría GA, García-Tojal J, Pis-Diez R, León IE, Parajón-Costa BP, González-Baró AC. Synthesis, Crystal Structure, Spectroscopic Characterization, DFT Calculations and Cytotoxicity Assays of a New Cu(II) Complex with an Acylhydrazone Ligand Derived from Thiophene. Inorganics. 2021; 9(2):9. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics9020009
Chicago/Turabian StyleRodríguez, María R., Lucía M. Balsa, Oscar E. Piro, Gustavo A. Etcheverría, Javier García-Tojal, Reinaldo Pis-Diez, Ignacio E. León, Beatriz P. Parajón-Costa, and Ana C. González-Baró. 2021. "Synthesis, Crystal Structure, Spectroscopic Characterization, DFT Calculations and Cytotoxicity Assays of a New Cu(II) Complex with an Acylhydrazone Ligand Derived from Thiophene" Inorganics 9, no. 2: 9. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics9020009