Study of DNA Interaction and Cytotoxicity Activity of Oxidovanadium(V) Complexes with ONO Donor Schiff Base Ligands
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis
2.2. Spectral Characteristics
2.2.1. IR Spectroscopy
2.2.2. Electronic Spectra
2.2.3. NMR Spectra
2.2.4. ESI Mass Spectra
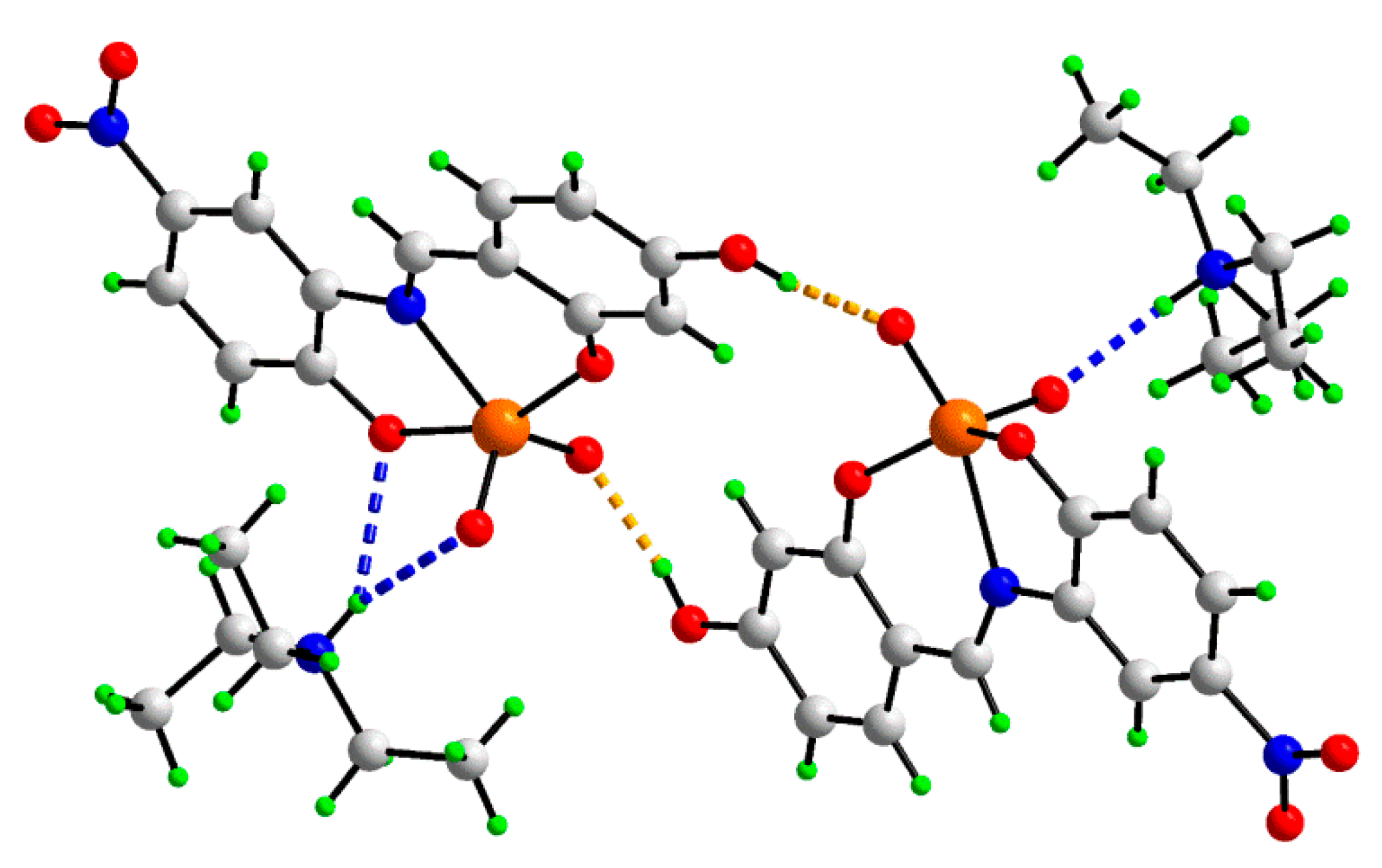
2.3. Single-Crystal X-ray Crystallography of 1
2.4. DNA-Binding Studies
2.4.1. UV–Vis Absorption Studies
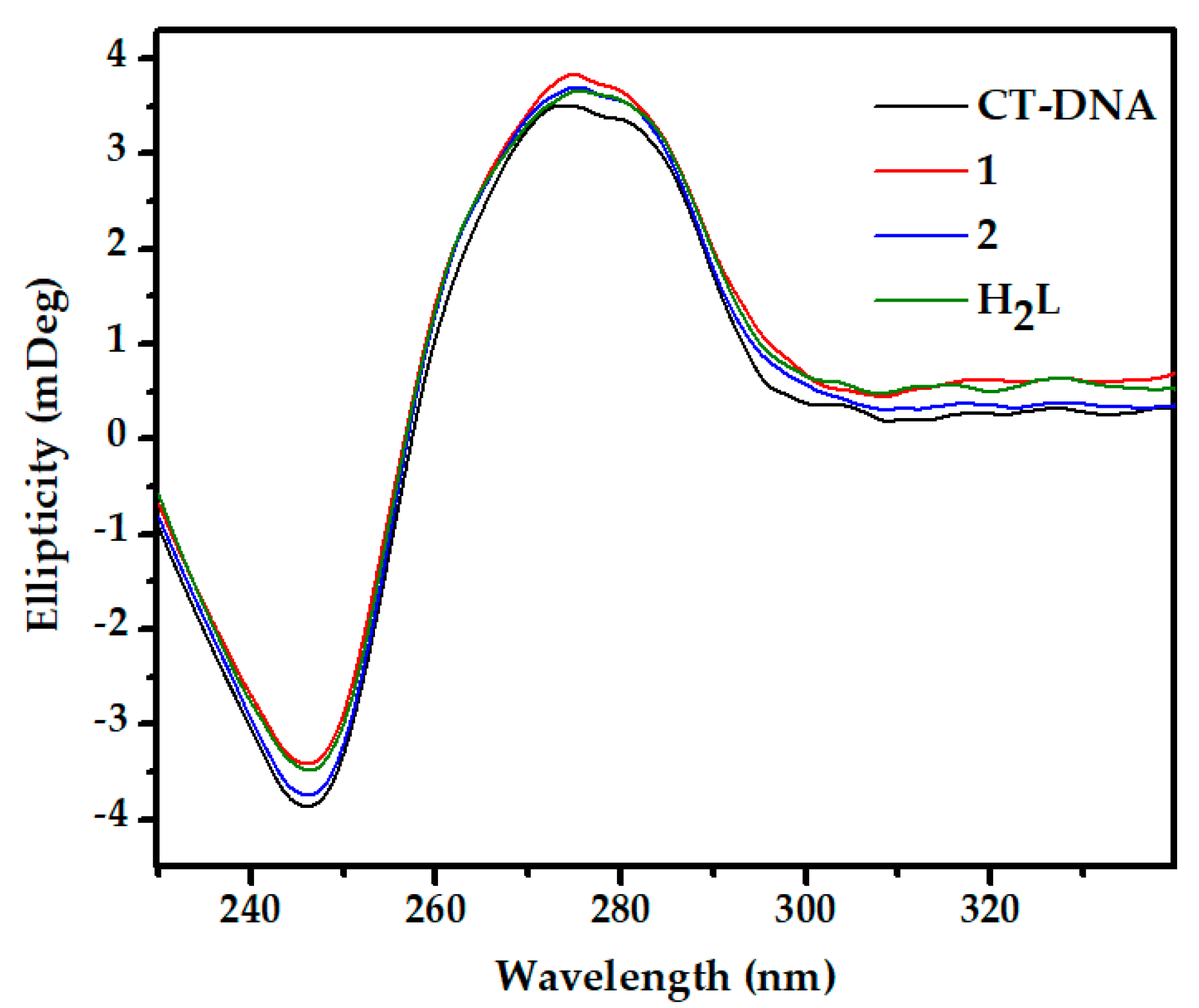
2.4.2. Circular Dichroism Studies
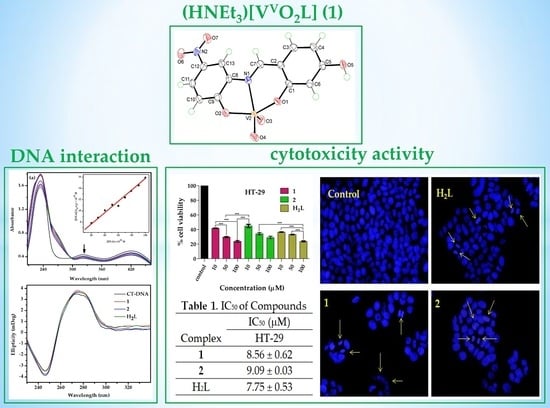
2.5. Cytotoxicity
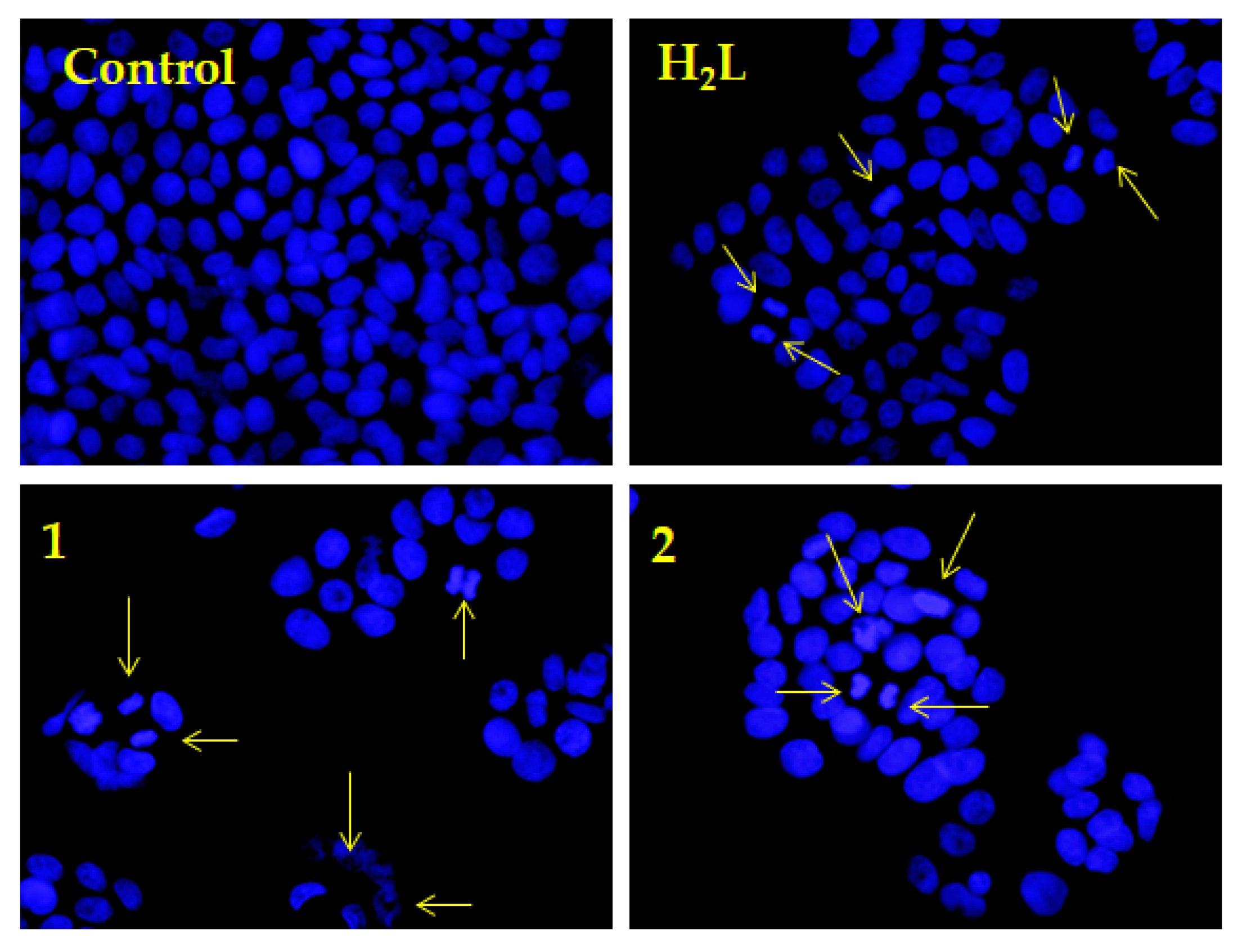
2.6. Nuclear DAPI Staining Assay
3. Materials and Methods
3.1. Materials
3.2. Synthesis of Ligands
3.3. Synthesis of Oxidovanadium(V) Complexes
3.4. Single-Crystal X-ray Crystallography
3.5. DNA-Binding Experiments
3.5.1. UV–Vis Absorption Studies
3.5.2. Circular Dichroism Studies
3.6. Cytotoxicity Analysis through MTT Assay
3.7. Nuclear DAPI Staining Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pessoa, J.C.; Garribba, E.; Santos, M.F.A.; Santos-Silva, T. Vanadium and proteins: Uptake, transport, structure, activity and function. Coord. Chem. Rev. 2015, 301–302, 49–86. [Google Scholar] [CrossRef]
- Rehder, D. The future of/for vanadium. Dalton Trans. 2013, 42, 11749–11761. [Google Scholar] [CrossRef]
- Thompson, K. Coordination chemistry of vanadium in metallopharmaceutical candidate compounds. Coord. Chem. Rev. 2001, 219–221, 1033–1053. [Google Scholar] [CrossRef]
- Baran, E.J. Oxovanadium(IV) and oxovanadium(V) complexes relevant to biological systems. J. Inorg. Biochem. 2000, 80, 1–10. [Google Scholar] [CrossRef]
- Domingues, N.; Pelletier, J.; Ostenson, C.-G.; Castro, M.M.C.A. Therapeutic properties of VO(dmpp)2 as assessed by in vitro and in vivo studies in type 2 diabetic GK rats. J. Inorg. Biochem. 2014, 131, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Pillai, S.I.; Subramanian, S.P.; Kandaswamy, M. A novel insulin mimetic vanadium-flavonol complex: Synthesis, characterization and in vivo evaluation in STZ-induced rats. Eur. J. Med. Chem. 2013, 63, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Turtoi, M.; Anghelache, M.; Patrascu, A.A.; Maxim, C.; Manduteanu, I.; Calin, M.; Popescu, D.-L. Synthesis, Characterization, and In Vitro Insulin-Mimetic Activity Evaluation of Valine Schiff Base Coordination Compounds of Oxidovanadium(V). Biomedicines 2021, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.P.; Pasayat, S.; Bhakat, S.; Roy, S.; Dinda, R.; Tiekink, E.R.T.; Mukhopadhyay, S.; Bhutia, S.K.; Hardikar, M.R.; Joshi, B.N.; et al. Highly stable hexacoordinated nonoxidovanadium(IV) complexes of sterically constrained ligands: Syntheses, structure, and study of antiproliferative and insulin mimetic activity. Inorg. Chem. 2013, 52, 14096–14107. [Google Scholar] [CrossRef] [PubMed]
- Orvig, C.; Caravan, P.; Gelmini, L.; Glover, N.; Herring, F.G.; Li, H.; McNeill, J.H.; Rettig, S.J.; Setyawati, I.A. Reaction chemistry of BMOV, bis(maltolato)oxovanadium(IV), a potent insulin mimetic agent. J. Am. Chem. Soc. 1995, 117, 12759–12770. [Google Scholar] [CrossRef]
- Evangelou, A.M. Vanadium in cancer treatment. Crit. Rev. Oncol. 2002, 42, 249–265. [Google Scholar] [CrossRef]
- Ebrahimipour, S.Y.; Sheikhshoaie, I.; Kautz, A.C.; Ameri, M.; Pasban-Aliabadi, H.; Amiri Rudbari, H.; Bruno, G.; Janiak, C. Mono- and dioxido-vanadium(V) complexes of a tridentate ONO Schiff base ligand: Synthesis, spectral characterization, X-ray crystal structure, and anticancer activity. Polyhedron 2015, 93, 99–105. [Google Scholar] [CrossRef]
- Levina, A.; Pires Vieira, A.; Wijetunga, A.; Kaur, R.; Koehn, J.T.; Crans, D.C.; Lay, P.A. A Short-Lived but Highly Cytotoxic Vanadium(V) Complex as a Potential Drug Lead for Brain Cancer Treatment by Intratumoral Injections. Angew. Chem. Int. Ed. Engl. 2020, 59, 15834–15838. [Google Scholar] [CrossRef]
- Banerjee, A.; Dash, S.P.; Mohanty, M.; Sahu, G.; Sciortino, G.; Garribba, E.; Carvalho, M.F.N.N.; Marques, F.; Costa Pessoa, J.; Kaminsky, W.; et al. New VIV, VIVO, VVO, and VVO2 Systems: Exploring their Interconversion in Solution, Protein Interactions, and Cytotoxicity. Inorg. Chem. 2020, 59, 14042–14057. [Google Scholar] [CrossRef]
- Bishayee, A.; Waghray, A.; Patel, M.A.; Chatterjee, M. Vanadium in the detection, prevention and treatment of cancer: The in vivo evidence. Cancer Lett. 2010, 294, 1–12. [Google Scholar] [CrossRef]
- Ni, L.; Zhao, H.; Tao, L.; Li, X.; Zhou, Z.; Sun, Y.; Chen, C.; Wei, D.; Liu, Y.; Diao, G. Synthesis, in vitro cytotoxicity, and structure-activity relationships (SAR) of multidentate oxidovanadium(IV) complexes as anticancer agents. Dalton Trans. 2018, 47, 10035–10045. [Google Scholar] [CrossRef] [PubMed]
- Sutradhar, M.; Pombeiro, A.J.L. Coordination chemistry of non-oxido, oxido and dioxidovanadium(IV/V) complexes with azine fragment ligands. Coord. Chem. Rev. 2014, 265, 89–124. [Google Scholar] [CrossRef]
- Rezaeivala, M.; Keypour, H. Schiff base and non-Schiff base macrocyclic ligands and complexes incorporating the pyridine moiety—The first 50 years. Coord. Chem. Rev. 2014, 280, 203–253. [Google Scholar] [CrossRef]
- Gupta, K.C.; Sutar, A.K. Catalytic activities of Schiff base transition metal complexes. Coord. Chem. Rev. 2008, 252, 1420–1450. [Google Scholar] [CrossRef]
- Liu, X.; Hamon, J.-R. Recent developments in penta-, hexa- and heptadentate Schiff base ligands and their metal complexes. Coord. Chem. Rev. 2019, 389, 94–118. [Google Scholar] [CrossRef]
- Khan, M.I.; Khan, A.; Hussain, I.; Khan, M.A.; Gul, S.; Iqbal, M.; Inayat-Ur-Rahman; Khuda, F. Spectral, XRD, SEM and biological properties of new mononuclear Schiff base transition metal complexes. Inorg. Chem. Commun. 2013, 35, 104–109. [Google Scholar] [CrossRef]
- Zhang, N.; Fan, Y.-h.; Zhang, Z.; Zuo, J.; Zhang, P.-f.; Wang, Q.; Liu, S.-B.; Bi, C.-F. Syntheses, crystal structures and anticancer activities of three novel transition metal complexes with Schiff base derived from 2-acetylpyridine and l-tryptophan. Inorg. Chem. Commun. 2012, 22, 68–72. [Google Scholar] [CrossRef]
- Tsuchida, E. Oxovanadium(III–V) mononuclear complexes and their linear assemblies bearing tetradentate Schiff base ligands: Structure and reactivity as multielectron redox catalysts. Coord. Chem. Rev. 2003, 237, 213–228. [Google Scholar] [CrossRef]
- Patra, S.; Chatterjee, S.; Si, T.K.; Mukherjea, K.K. Synthesis, structural characterization, VHPO mimicking peroxidative bromination and DNA nuclease activity of oxovanadium(V) complexes. Dalton Trans. 2013, 42, 13425–13435. [Google Scholar] [CrossRef]
- Sathyadevi, P.; Krishnamoorthy, P.; Butorac, R.R.; Cowley, A.H.; Bhuvanesh, N.S.P.; Dharmaraj, N. Effect of substitution and planarity of the ligand on DNA/BSA interaction, free radical scavenging and cytotoxicity of diamagnetic Ni(II) complexes: A systematic investigation. Dalton Trans. 2011, 40, 9690–9702. [Google Scholar] [CrossRef] [Green Version]
- Raj Kumar, R.; Mohamed Subarkhan, M.K.; Ramesh, R. Synthesis and structure of nickel(II) thiocarboxamide complexes: Effect of ligand substitutions on DNA/protein binding, antioxidant and cytotoxicity. RSC Adv. 2015, 5, 46760–46773. [Google Scholar] [CrossRef]
- Galati, G.; Chan, T.; Wu, B.; O’Brien, P.J. Glutathione-dependent generation of reactive oxygen species by the peroxidase-catalyzed redox cycling of flavonoids. Chem. Res. Toxicol. 1999, 12, 521–525. [Google Scholar] [CrossRef]
- Galati, G. Prooxidant activity and cellular effects of the phenoxyl radicals of dietary flavonoids and other polyphenolics. Toxicology 2002, 177, 91–104. [Google Scholar] [CrossRef]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Prior, R.L.; Wu, X.; Gu, L. Flavonoid metabolism and challenges to understanding mechanisms of health effects. J. Sci. Food Agric. 2006, 86, 2487–2491. [Google Scholar] [CrossRef]
- Kessler, M.; Ubeaud, G.; Jung, L. Anti- and pro-oxidant activity of rutin and quercetin derivatives. J. Pharm. Pharmacol. 2003, 55, 131–142. [Google Scholar] [CrossRef]
- Pasayat, S.; Böhme, M.; Dhaka, S.; Dash, S.P.; Majumder, S.; Maurya, M.R.; Plass, W.; Kaminsky, W.; Dinda, R. Synthesis, Theoretical Study and Catalytic Application of Oxidometal (Mo or V) Complexes: Unexpected Coordination Due to Ligand Rearrangement through Metal-Mediated C-C Bond Formation. Eur. J. Inorg. Chem. 2016, 2016, 1604–1618. [Google Scholar] [CrossRef]
- Dinda, R.; Majhi, P.K.; Sengupta, P.; Pasayat, S.; Ghosh, S.; Falvello, L.R.; Mak, T.C.W. Alkali metal (Na+ and K+)-mediated supramolecular assembly of oxovanadium(V) complexes: Synthesis and structural characterization. Polyhedron 2010, 29, 248–253. [Google Scholar] [CrossRef] [Green Version]
- Dash, S.P.; Pasayat, S.; Saswati; Dash, H.R.; Das, S.; Butcher, R.J.; Dinda, R. Oxovanadium(V) complexes incorporating tridentate aroylhydrazoneoximes: Synthesis, characterizations and antibacterial activity. Polyhedron 2012, 31, 524–529. [Google Scholar] [CrossRef]
- Dash, S.P.; Panda, A.K.; Dhaka, S.; Pasayat, S.; Biswas, A.; Maurya, M.R.; Majhi, P.K.; Crochet, A.; Dinda, R. A study of DNA/BSA interaction and catalytic potential of oxidovanadium(V) complexes with ONO donor ligands. Dalton Trans. 2016, 45, 18292–18307. [Google Scholar] [CrossRef] [Green Version]
- Saswati; Adão, P.; Majumder, S.; Dash, S.P.; Roy, S.; Kuznetsov, M.L.; Pessoa, J.C.; Gomes, C.S.B.; Hardikar, M.R.; Tiekink, E.R.T.; et al. Synthesis, structure, solution behavior, reactivity and biological evaluation of oxidovanadium(IV/V) thiosemicarbazone complexes. Dalton Trans. 2018, 47, 11358–11374. [Google Scholar] [CrossRef]
- Banerjee, A.; Dash, S.P.; Mohanty, M.; Sanna, D.; Sciortino, G.; Ugone, V.; Garribba, E.; Reuter, H.; Kaminsky, W.; Dinda, R. Chemistry of mixed-ligand oxidovanadium(IV) complexes of aroylhydrazones incorporating quinoline derivatives: Study of solution behavior, theoretical evaluation and protein/DNA interaction. J. Inorg. Biochem. 2019, 199, 110786. [Google Scholar] [CrossRef]
- Roy, S.; Böhme, M.; Dash, S.P.; Mohanty, M.; Buchholz, A.; Plass, W.; Majumder, S.; Kulanthaivel, S.; Banerjee, I.; Reuter, H.; et al. Anionic Dinuclear Oxidovanadium(IV) Complexes with Azo Functionalized Tridentate Ligands and μ-Ethoxido Bridge Leading to an Unsymmetric Twisted Arrangement: Synthesis, X-ray Structure, Magnetic Properties, and Cytotoxicity. Inorg. Chem. 2018, 57, 5767–5781. [Google Scholar] [CrossRef]
- Dash, S.P.; Panda, A.K.; Pasayat, S.; Dinda, R.; Biswas, A.; Tiekink, E.R.T.; Mukhopadhyay, S.; Bhutia, S.K.; Kaminsky, W.; Sinn, E. Oxidovanadium(V) complexes of aroylhydrazones incorporating heterocycles: Synthesis, characterization and study of DNA binding, photo-induced DNA cleavage and cytotoxic activities. RSC Adv. 2015, 5, 51852–51867. [Google Scholar] [CrossRef]
- Lima, S.; Banerjee, A.; Mohanty, M.; Sahu, G.; Kausar, C.; Patra, S.K.; Garribba, E.; Kaminsky, W.; Dinda, R. Synthesis, structure and biological evaluation of mixed ligand oxidovanadium(IV ) complexes incorporating 2-(arylazo)phenolates. New J. Chem. 2019, 43, 17711–17725. [Google Scholar] [CrossRef]
- Banerjee, A.; Mohanty, M.; Lima, S.; Samanta, R.; Garribba, E.; Sasamori, T.; Dinda, R. Synthesis, structure and characterization of new dithiocarbazate-based mixed ligand oxidovanadium(IV ) complexes: DNA/HSA interaction, cytotoxic activity and DFT studies. New J. Chem. 2020, 44, 10946–10963. [Google Scholar] [CrossRef]
- Dash, S.P.; Panda, A.K.; Pasayat, S.; Majumder, S.; Biswas, A.; Kaminsky, W.; Mukhopadhyay, S.; Bhutia, S.K.; Dinda, R. Evaluation of the cell cytotoxicity and DNA/BSA binding and cleavage activity of some dioxidovanadium(V) complexes containing aroylhydrazones. J. Inorg. Biochem. 2015, 144, 1–12. [Google Scholar] [CrossRef]
- Mohanty, M.; Maurya, S.K.; Banerjee, A.; Patra, S.A.; Maurya, M.R.; Crochet, A.; Brzezinski, K.; Dinda, R. In vitro cytotoxicity and catalytic evaluation of dioxidovanadium(V) complexes in an azohydrazone ligand environment. New J. Chem. 2019, 43, 17680–17695. [Google Scholar] [CrossRef] [Green Version]
- Dash, S.P.; Panda, A.K.; Pasayat, S.; Dinda, R.; Biswas, A.; Tiekink, E.R.T.; Patil, Y.P.; Nethaji, M.; Kaminsky, W.; Mukhopadhyay, S.; et al. Syntheses and structural investigation of some alkali metal ion-mediated LVVO2− (L2− = tridentate ONO ligands) species: DNA binding, photo-induced DNA cleavage and cytotoxic activities. Dalton Trans. 2014, 43, 10139–10156. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.P.; Roy, S.; Mohanty, M.; Carvalho, M.F.N.N.; Kuznetsov, M.L.; Pessoa, J.C.; Kumar, A.; Patil, Y.P.; Crochet, A.; Dinda, R. Versatile Reactivity and Theoretical Evaluation of Mono- and Dinuclear Oxidovanadium(V) Compounds of Aroylazines: Electrogeneration of Mixed-Valence Divanadium(IV,V) Complexes. Inorg. Chem. 2016, 55, 8407–8421. [Google Scholar] [CrossRef] [Green Version]
- Dash, S.P.; Majumder, S.; Banerjee, A.; Carvalho, M.F.N.N.; Adão, P.; Pessoa, J.C.; Brzezinski, K.; Garribba, E.; Reuter, H.; Dinda, R. Chemistry of Monomeric and Dinuclear Non-Oxido Vanadium(IV) and Oxidovanadium(V) Aroylazine Complexes: Exploring Solution Behavior. Inorg. Chem. 2016, 55, 1165–1182. [Google Scholar] [CrossRef] [PubMed]
- Nemeikaite-Ceniene, A.; Imbrasaite, A.; Sergediene, E.; Cenas, N. Quantitative structure-activity relationships in prooxidant cytotoxicity of polyphenols: Role of potential of phenoxyl radical/phenol redox couple. Arch. Biochem. Biophys. 2005, 441, 182–190. [Google Scholar] [CrossRef]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, G.C. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 1349–1356. [Google Scholar] [CrossRef]
- Spek, A.L. checkCIF validation ALERTS: What they mean and how to respond. Acta Crystallogr. E Crystallogr. Commun. 2020, 76, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Krishnamoorthy, P.; Sathyadevi, P.; Butorac, R.R.; Cowley, A.H.; Bhuvanesh, N.S.P.; Dharmaraj, N. Copper(I) and nickel(II) complexes with 1:1 vs. 1:2 coordination of ferrocenyl hydrazone ligands: Do the geometry and composition of complexes affect DNA binding/cleavage, protein binding, antioxidant and cytotoxic activities? Dalton Trans. 2012, 41, 4423–4436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirajuddin, M.; Ali, S.; Badshah, A. Drug-DNA interactions and their study by UV-Visible, fluorescence spectroscopies and cyclic voltametry. J. Photochem. Photobiol. B 2013, 124, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Saswati; Mohanty, M.; Banerjee, A.; Biswal, S.; Horn, A.; Schenk, G.; Brzezinski, K.; Sinn, E.; Reuter, H.; Dinda, R. Polynuclear zinc(II) complexes of thiosemicarbazone: Synthesis, X-ray structure and biological evaluation. J. Inorg. Biochem. 2020, 203, 110908. [Google Scholar] [CrossRef] [PubMed]
- Ganeshpandian, M.; Loganathan, R.; Suresh, E.; Riyasdeen, A.; Akbarsha, M.A.; Palaniandavar, M. New ruthenium(II) arene complexes of anthracenyl-appended diazacycloalkanes: Effect of ligand intercalation and hydrophobicity on DNA and protein binding and cleavage and cytotoxicity. Dalton Trans. 2014, 43, 1203–1219. [Google Scholar] [CrossRef]
- Tan, J.; Wang, B.; Zhu, L. DNA binding and oxidative DNA damage induced by a quercetin copper(II) complex: Potential mechanism of its antitumor properties. J. Biol. Inorg. Chem. 2009, 14, 727–739. [Google Scholar] [CrossRef]
- Perron, N.R.; Hodges, J.N.; Jenkins, M.; Brumaghim, J.L. Predicting how polyphenol antioxidants prevent DNA damage by binding to iron. Inorg. Chem. 2008, 47, 6153–6161. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Krishnamoorthy, P.; Sathyadevi, P.; Cowley, A.H.; Butorac, R.R.; Dharmaraj, N. Evaluation of DNA binding, DNA cleavage, protein binding and in vitro cytotoxic activities of bivalent transition metal hydrazone complexes. Eur. J. Med. Chem. 2011, 46, 3376–3387. [Google Scholar] [CrossRef]
- Raja, D.S.; Paramaguru, G.; Bhuvanesh, N.S.P.; Reibenspies, J.H.; Renganathan, R.; Natarajan, K. Effect of terminal N-substitution in 2-oxo-1,2-dihydroquinoline-3-carbaldehyde thiosemicarbazones on the mode of coordination, structure, interaction with protein, radical scavenging and cytotoxic activity of copper(II) complexes. Dalton Trans. 2011, 40, 4548–4559. [Google Scholar] [CrossRef]
- Chen, Y.; Qin, M.-Y.; Wu, J.-H.; Wang, L.; Chao, H.; Ji, L.-N.; Xu, A.-L. Synthesis, characterization, and anticancer activity of ruthenium(II)-β-carboline complex. Eur. J. Med. Chem. 2013, 70, 120–129. [Google Scholar] [CrossRef]
- Mohamadi, M.; Yousef Ebrahimipour, S.; Torkzadeh-Mahani, M.; Foro, S.; Akbari, A. A mononuclear diketone-based oxido-vanadium(V) complex: Structure, DNA and BSA binding, molecular docking and anticancer activities against MCF-7, HPG-2, and HT-29 cell lines. RSC Adv. 2015, 5, 101063–101075. [Google Scholar] [CrossRef]
- Moeller, T. (Ed.) Inorganic Syntheses; Mc-Graw Hill Book Company: New York, NY, USA, 1957; Volume 5, pp. 113–116. [Google Scholar]
- Rigaku Oxford Diffraction. CrysAlisPRO; Rigaku Oxford Diffraction Ltd.: Oxfordshire, UK, 2017. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Brandenburg, K.; Berndt, M. DIAMOND, Version 3.2k; GbR: Bonn, Germany, 2006. [Google Scholar]
- Roy, S.; Mohanty, M.; Miller, R.G.; Patra, S.A.; Lima, S.; Banerjee, A.; Metzler-Nolte, N.; Sinn, E.; Kaminsky, W.; Dinda, R. Probing CO Generation through Metal-Assisted Alcohol Dehydrogenation in Metal-2-(arylazo)phenol Complexes Using Isotopic Labeling (Metal = Ru, Ir): Synthesis, Characterization, and Cytotoxicity Studies. Inorg. Chem. 2020, 59, 15526–15540. [Google Scholar] [CrossRef]
- Bhutia, S.K.; Mallick, S.K.; Maiti, S.; Maiti, T.K. Antitumor and proapoptotic effect of Abrus agglutinin derived peptide in Dalton’s lymphoma tumor model. Chem. Biol. Interact. 2008, 174, 11–18. [Google Scholar] [CrossRef]


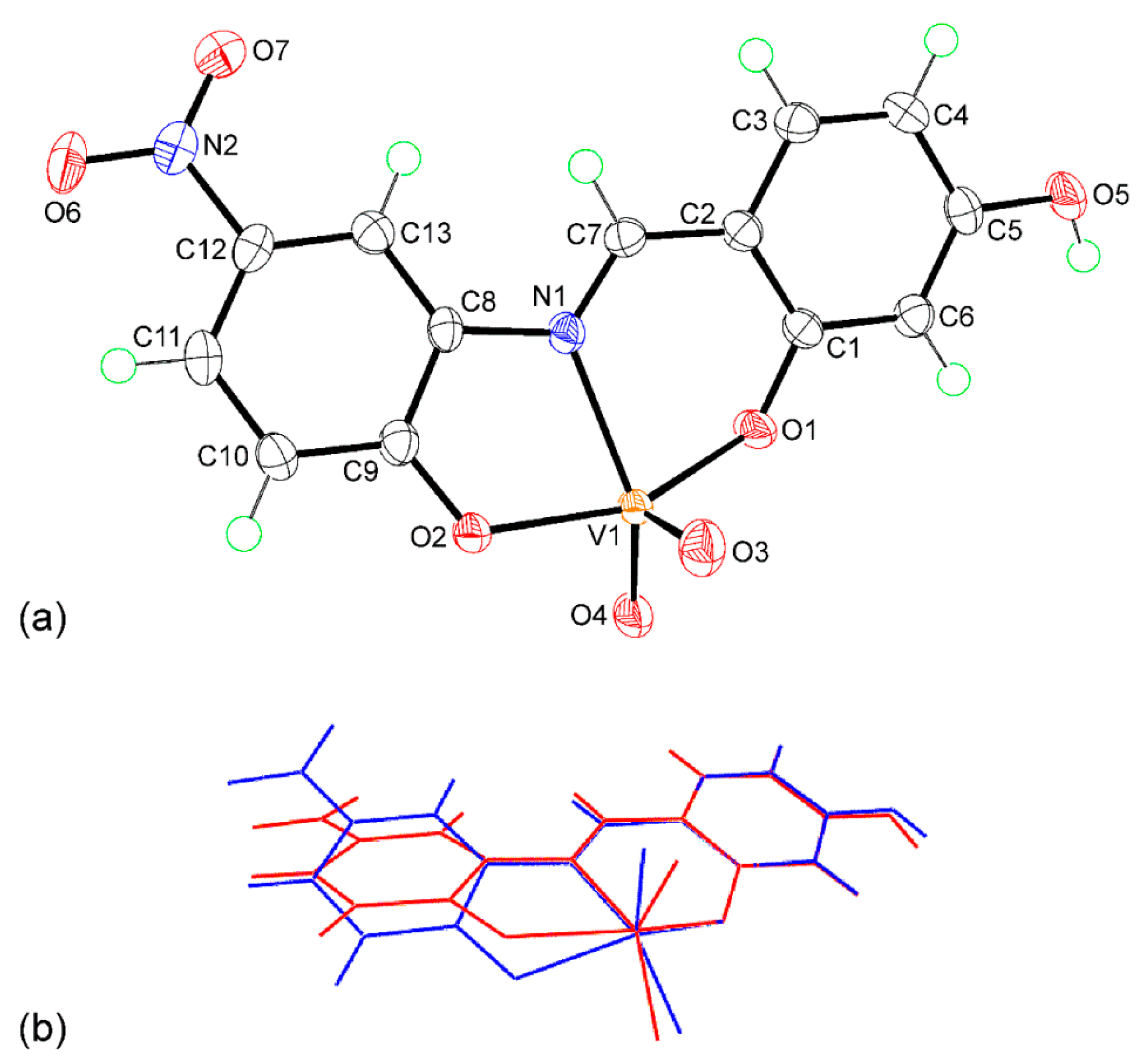


| Parameter | Anion “a” | Anion “b” |
|---|---|---|
| V–O1 | 1.9152(9) | 1.8942(9) |
| V–O2 | 1.9735(9) | 1.9191(9) |
| V–O3 | 1.6219(10) | 1.6356(10) |
| V–O4 | 1.6463(9) | 1.6473(10) |
| V–N1 | 2.1658(11) | 2.1968(11) |
| C7–N1 | 1.3033(17) | 1.2942(17) |
| O1–V–O2 | 155.41(4) | 140.66(4) |
| O3–V–O4 | 109.05(5) | 108.51(5) |
| N1–V–O4 | 140.02(5) | 155.17(5) |
| A | H | B | H...B | A...B | A–H...B |
|---|---|---|---|---|---|
| O5a | H1o | O4b | 1.825(12) | 2.6548(14) | 171.3(18) |
| O5b | H2o | O4a | 1.830(15) | 2.6640(14) | 171.7(17) |
| N3a | H1n | O2a | 2.466(13) | 3.2244(15) | 145.4(13) |
| N3a | H1n | O3a | 2.224(12) | 2.9473(15) | 139.8(14) |
| N3b | H2n | O3b | 1.918(12) | 2.7904(16) | 170.7(14) |
| Complex | Binding Constants (Kb) (M−1) |
|---|---|
| 1 | 2.81 × 104 |
| 2 | 2.35 × 104 |
| H2L | 1.59 × 104 |
| Compound | IC50 (μM) | |
|---|---|---|
| HT-29 | NIH-3T3 | |
| 1 | 8.56 ± 0.62 | 67.85 ± 5.48 |
| 2 | 9.09 ± 0.03 | 79.77 ± 4.00 |
| H2L | 7.75 ± 0.53 | 69.32 ± 4.42 |
| Formula | [C6H16N][C13H8N2O7V] |
|---|---|
| Molecular weight | 457.35 |
| Crystal system | triclinic |
| Space group | P1 |
| a/Å | 10.8226(1) |
| b/Å | 10.9399(2) |
| c/Å | 17.3389(3) |
| α/° | 79.412(1) |
| β/° | 78.905(1) |
| γ/° | 86.861(1) |
| V/Å3 | 1979.82(5) |
| Z | 4 |
| Dc/g cm−3 | 1.534 |
| μ/mm−1 | 4.621 |
| Measured data | 46,958 |
| Unique data | 7061 |
| Observed data (I ≥ 2.0σ(I)) | 6958 |
| No. parameters | 559 |
| R, obs. data; all data | 0.025; 0.026 |
| Rw, obs. data; all data | 0.072; 0.072 |
| Range of residual electron | |
| density peaks/eÅ−3 | −0.58–0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahu, G.; Tiekink, E.R.T.; Dinda, R. Study of DNA Interaction and Cytotoxicity Activity of Oxidovanadium(V) Complexes with ONO Donor Schiff Base Ligands. Inorganics 2021, 9, 66. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics9090066
Sahu G, Tiekink ERT, Dinda R. Study of DNA Interaction and Cytotoxicity Activity of Oxidovanadium(V) Complexes with ONO Donor Schiff Base Ligands. Inorganics. 2021; 9(9):66. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics9090066
Chicago/Turabian StyleSahu, Gurunath, Edward R. T. Tiekink, and Rupam Dinda. 2021. "Study of DNA Interaction and Cytotoxicity Activity of Oxidovanadium(V) Complexes with ONO Donor Schiff Base Ligands" Inorganics 9, no. 9: 66. https://0-doi-org.brum.beds.ac.uk/10.3390/inorganics9090066