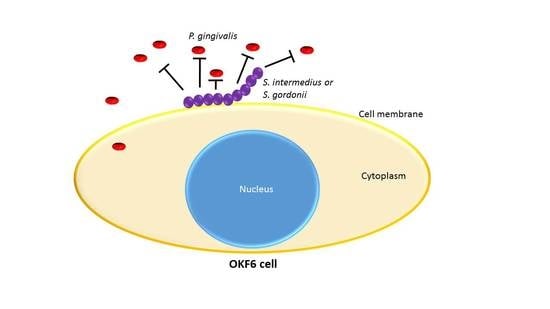
Effects of Oral Commensal Streptococci on Porphyromonas gingivalis Invasion into Oral Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Bacterial Cultures
2.2. Bacterial Growth Curves
2.3. Streptococcal Cytotoxicity Assay
2.4. Streptococcal Adherence and Invasion Assays
2.5. P. gingivalis Invasion Assays
2.6. Streptococcal Inhibition of P. gingivalis Growth
2.7. Statistical Analyses
3. Results
3.1. KSFM Supports Growth of Commensal Bacteria
3.2. OKF6 Cell Viability after Commensal Challenge
3.3. Commensal Bacteria Adherence Assay
3.4. P. gingivalis Invasion Post-Commensal Co-Cultures
3.5. Commensal Antagonistic Properties against P. gingivalis
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Periodontal Disease Division of Oral Health CDC. Available online: https://www.cdc.gov/oralhealth/periodontal_disease/index.htm (accessed on 23 May 2018).
- Eke, P.I.; Wei, L.; Borgnakke, W.; Zhang, X.; McGuire, L.C.; Genco, R.J.; Thornton-Evans, G.; Lu, H. Periodontitis prevalence in adults? 65 years of age, in the USA. Periodontology 2000 2016, 72, 76–95. [Google Scholar] [CrossRef] [PubMed]
- Periodontitis: MedlinePlus Medical Encyclopedia. Available online: https://medlineplus.gov/ency/article/001059.htm (accessed on 23 May 2018).
- Parahitiyawa, N.B.; Jin, L.J.; Leung, W.; Yam, W.C.; Samaranayake, L.P. Microbiology of Odontogenic Bacteremia: Beyond Endocarditis. Clin. Microbiol. Rev. 2009, 22, 46–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P. Oral microbiota and systemic disease. Anaerobe 2013, 24, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.; Ford, P.J.; Cullinan, M.; Leishman, S.; Yamazaki, K. Relationship between periodontal infections and systemic disease. Clin. Microbiol. Infect. 2007, 13, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Han, J.; Liu, Z.; Song, Y.; Wang, Z.; Sun, Z. Effects of periodontal treatment on lung function and exacerbation frequency in patients with chronic obstructive pulmonary disease and chronic periodontitis: A 2-year pilot randomized controlled trial. J. Clin. Periodontol. 2014, 41, 564–572. [Google Scholar] [CrossRef]
- Wu, Z.; Nakanishi, H. Lessons from Microglia Aging for the Link between Inflammatory Bone Disorders and Alzheimer’s Disease. Available online: https://www.hindawi.com/journals/jir/2015/471342/ (accessed on 12 June 2018).
- Scannapieco, F. The oral microbiome: Its role in health and in oral and systemic infections. Clin. Microbiol. Newsl. 2013, 35, 163–169. [Google Scholar] [CrossRef]
- Borgnakke, W.; Ylöstalo, P.V.; Taylor, G.W.; Genco, R.J. Effect of periodontal disease on diabetes: Systematic review of epidemiologic observational evidence. J. Clin. Periodontol. 2013, 40, 135–152. [Google Scholar] [CrossRef]
- Gaur, S.; Agnihotri, R. Alzheimer’s disease and chronic periodontitis: Is there an association? Geriatr. Gerontol. Int. 2014, 15, 391–404. [Google Scholar] [CrossRef]
- Singhrao, S.K.; Harding, A.; Poole, S.; Kesavalu, L.; Crean, S. Porphyromonas Gingivalis Periodontal Infection and Its Putative Links with Alzheimer’s Disease. Available online: https://www.hindawi.com/journals/mi/2015/137357/ (accessed on 12 June 2018).
- Ho, M.-H.; Lamont, R.J.; Xie, H. Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis. Sci. Rep. 2017, 7, 1413. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Liang, S.; Payne, M.A.; Hashim, A.; Jotwani, R.; Eskan, M.A.; McIntosh, M.L.; Alsam, A.; Kirkwood, K.L.; Lambris, J.D.; et al. Low-Abundance Biofilm Species Orchestrates Inflammatory Periodontal Disease through the Commensal Microbiota and Complement. Cell Host Microbe 2011, 10, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Tribble, G.; Lamont, R.J. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontology 2000 2010, 52, 68–83. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, T.; Kraus, D.; Probstmeier, R.; Jepsen, S.; Winter, J. Stimulation with Porphyromonas gingivalis enhances malignancy and initiates anoikis resistance in immortalized oral keratinocytes. J. Cell. Physiol. 2019, 234, 21903–21914. [Google Scholar] [CrossRef]
- Lamont, R.J.; El-Sabaeny, A.; Park, Y.; Cook, G.S.; Costerton, J.W.; DeMuth, D.R. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 2002, 148, 1627–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.; Simionato, M.R.; Sekiya, K.; Murakami, Y.; James, D.; Chen, W.; Hackett, M.; Yoshimura, F.; DeMuth, D.R.; Lamont, R. Short Fimbriae of Porphyromonas gingivalis and Their Role in Coadhesion with Streptococcus gordonii. Infect. Immun. 2005, 73, 3983–3989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutt, B.; Ren, Q.; Hauser-Gerspach, I.; Kardas, P.; Stübinger, S.; Astasov-Frauenhoffer, M.; Waltimo, T. Beneficial Oral Biofilms as Smart Bioactive Interfaces. Front. Microbiol. 2018, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrero, E.R.; Slomka, V.; Bernaerts, K.; Boon, N.; Hernandez-Sanabria, E.; Passoni, B.B.; Quirynen, M.; Teughels, W. Antimicrobial effects of commensal oral species are regulated by environmental factors. J. Dent. 2016, 47, 23–33. [Google Scholar] [CrossRef]
- Handfield, M.; Mans, J.J.; Zheng, G.; Lopez, M.C.; Mao, S.; Progulske-Fox, A.; Narasimhan, G.; Baker, H.V.; Lamont, R.J. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell. Microbiol. 2005, 7, 811–823. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Mans, J.J.; Mao, S.; Lopez, M.C.; Baker, H.V.; Handfield, M.; Lamont, R.J. Gingival Epithelial Cell Transcriptional Responses to Commensal and Opportunistic Oral Microbial Species. Infect. Immun. 2007, 75, 2540–2547. [Google Scholar] [CrossRef] [Green Version]
- Rosan, B.; Lamont, R.J. Dental plaque formation. Microbes Infect. 2000, 2, 1599–1607. [Google Scholar] [CrossRef]
- Facklam, R. What Happened to the Streptococci: Overview of Taxonomic and Nomenclature Changes. Clin. Microbiol. Rev. 2002, 15, 613–630. [Google Scholar] [CrossRef] [Green Version]
- Colombo, A.V.; Da Silva, C.M.; Haffajee, A.; Colombo, A.P.V. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J. Periodontal Res. 2007, 42, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.C.; Moffatt, C.E.; Hagerty, D.; Whitmore, S.E.; A Brown, T.; Graves, D.T.; Lamont, R.J. Interaction of oral bacteria with gingival epithelial cell multilayers. Mol. Oral Microbiol. 2011, 26, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Roberts, J.S.; Choi, C.H.; Atanasova, K.; Yilmaz, Ö. Porphyromonas gingivalis traffics into endoplasmic reticulum-rich-autophagosomes for successful survival in human gingival epithelial cells. Virulence 2018, 9, 845–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamont, R.J.; Chan, A.; Belton, C.M.; Izutsu, K.T.; Vasel, D.; Weinberg, A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 1995, 63, 3878–3885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, Ö.; Watanabe, K.; Lamont, R.J. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell. Microbiol. 2002, 4, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, I.; Amano, A.; Kuboniwa, M.; Nakamura, T.; Kawabata, S.; Hamada, S. Functional Differences among FimA Variants of Porphyromonas gingivalis and Their Effects on Adhesion to and Invasion of Human Epithelial Cells. Infect. Immun. 2002, 70, 277–285. [Google Scholar] [CrossRef] [Green Version]
- Cugini, C.; Stephens, D.N.; Nguyen, D.; Kantarci, A.; Davey, M.E. Arginine deiminase inhibits Porphyromonas gingivalis surface attachment. Microbiology 2013, 159, 275–285. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Hong, J.; Sharma, A.; Wang, B. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J. Periodontal Res. 2012, 47, 578–583. [Google Scholar] [CrossRef] [Green Version]
- Christopher, A.B.; Arndt, A.; Cugini, C.; Davey, M.E. A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development. Microbiology 2010, 156, 3469–3477. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.-Y.; Wu, J.; Lamont, R.J.; Lin, X.; Xie, H. Negative Correlation of Distributions of Streptococcus cristatus and Porphyromonas gingivalis in Subgingival Plaque. J. Clin. Microbiol. 2009, 47, 3902–3906. [Google Scholar] [CrossRef] [Green Version]
- Kreth, J.; Zhang, Y.; Herzberg, M.C. Streptococcal Antagonism in Oral Biofilms: Streptococcus sanguinis and Streptococcus gordonii Interference with Streptococcus mutans. J. Bacteriol. 2008, 190, 4632–4640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.-Y.; Kuramitsu, H.K. Interactions between Oral Bacteria: Inhibition of Streptococcus mutans Bacteriocin Production by Streptococcus gordonii. Appl. Environ. Microbiol. 2005, 71, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Sukeno, A.; Nagamune, H.; Whiley, R.A.; Jafar, S.I.; Opoku, A.; Ohkura, K.; Maeda, T.; Hirota, K.; Miyake, Y.; Kourai, H. Intermedilysin Is Essential for the Invasion of Hepatoma HepG2 Cells byStreptococcus intermedius. Microbiol. Immunol. 2005, 49, 681–694. [Google Scholar] [CrossRef]
- Nagata, E.; De Toledo, A.; Oho, T. Invasion of human aortic endothelial cells by oral viridans group streptococci and induction of inflammatory cytokine production. Mol. Oral Microbiol. 2010, 26, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Ye, P.; Harty, D.; Commandeur, Z.; Hunter, N. Binding of Streptococcus gordonii to oral epithelial monolayers increases paracellular barrier function. Microb. Pathog. 2013, 56, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Brice, D.C.; Figgins, E.; Yu, F.; Diamond, G. Type I interferon and interferon-stimulated gene expression in oral epithelial cells. Mol. Oral Microbiol. 2019, 34, 245–253. [Google Scholar] [CrossRef]
- Ho, M.-H.; Lamont, R.J.; Xie, H. A novel peptidic inhibitor derived from Streptococcus cristatus ArcA attenuates virulence potential of Porphyromonas gingivalis. Sci. Rep. 2017, 7, 16217. [Google Scholar] [CrossRef] [Green Version]
- Saito, A.; Inagaki, S.; Ishihara, K. Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis. Microb. Pathog. 2009, 47, 329–333. [Google Scholar] [CrossRef] [Green Version]
- Jauregui, C.; Robinson, B.; De Moya, A.V.; Brockman, R.D.; Roman, A.V.; Cash, M.N.; Culp, D.J.; Lamont, R.J. Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J. Periodontal Res. 2013, 48, 713–721. [Google Scholar] [CrossRef] [Green Version]
- Dommisch, H.; Reinartz, M.; Backhaus, T.; Deschner, J.; Chung, W.; Jepsen, S. Antimicrobial responses of primary gingival cells to Porphyromonas gingivalis. J. Clin. Periodontol. 2012, 39, 913–922. [Google Scholar] [CrossRef]
- O’Callaghan, A.; Corr, S. Establishing Boundaries: The Relationship That Exists between Intestinal Epithelial Cells and Gut-Dwelling Bacteria. Microorganisms 2019, 7, 663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell. Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, R.L.; Sequeira, R.P.; Clarke, T.B. The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 2017, 8, 1512. [Google Scholar] [CrossRef] [PubMed]
- Huck, O.; Mulhall, H.; Rubin, G.; Kizelnik, Z.; Iyer, R.; Perpich, J.D.; Haque, N.; Cani, P.D.; De Vos, W.M.; Amar, S. Akkermansia muciniphila reduces Porphyromonas gingivalis -induced inflammation and periodontal bone destruction. J. Clin. Periodontol. 2019, 47, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Kim, Y.; Min, B.-M.; Han, S.H.; Choi, Y. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J. Periodontal Res. 2007, 42, 503–510. [Google Scholar] [CrossRef]
- Chung, W.O.; Dale, B.A. Innate Immune Response of Oral and Foreskin Keratinocytes: Utilization of Different Signaling Pathways by Various Bacterial Species. Infect. Immun. 2004, 72, 352–358. [Google Scholar] [CrossRef] [Green Version]
- Sliepen, I.; Van Damme, J.; Van Essche, M.; Loozen, G.; Quirynen, M.; Teughels, W. Microbial Interactions Influence Inflammatory Host Cell Responses. J. Dent. Res. 2009, 88, 1026–1030. [Google Scholar] [CrossRef]
- Peyyala, R.; Kirakodu, S.S.; Novak, K.F.; Ebersole, J.L. Oral epithelial cell responses to multispecies microbial biofilms. J. Dent. Res. 2013, 92, 235–240. [Google Scholar] [CrossRef]
- Stathopoulou, P.G.; Benakanakere, M.R.; Galicia, J.C.; Kinane, D.F. Epithelial cell pro-inflammatory cytokine response differs across dental plaque bacterial species. J. Clin. Periodontol. 2010, 37, 24–29. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Chen, R.; Rudney, J. Streptococcus cristatus attenuates Fusobacterium nucleatum-induced interleukin-8 expression in oral epithelial cells. J. Periodontal Res. 2008, 43, 408–416. [Google Scholar] [CrossRef]
- Lamont, R.J.; Jenkinson, H.F. Life Below the Gum Line: Pathogenic Mechanisms ofPorphyromonas gingivalis. Microbiol. Mol. Boil. Rev. 1998, 62, 1244–1263. [Google Scholar] [CrossRef] [Green Version]
- Daalderop, L.; Wieland, B.; Tomsin, K.; Reyes, L.; Kramer, B.; Vanterpool, S.; Been, J.V. Periodontal Disease and Pregnancy Outcomes: Overview of Systematic Reviews. JDR Clin. Transl. Res. 2017, 3, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Lamont, R.J. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmlund, A.; Lampa, E.; Lind, L. Oral health and cardiovascular disease risk in a cohort of periodontitis patients. Atherosclerosis 2017, 262, 101–106. [Google Scholar] [CrossRef] [PubMed]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanel, A.N.; Herzog, H.M.; James, M.G.; Cuadra, G.A. Effects of Oral Commensal Streptococci on Porphyromonas gingivalis Invasion into Oral Epithelial Cells. Dent. J. 2020, 8, 39. https://0-doi-org.brum.beds.ac.uk/10.3390/dj8020039
Hanel AN, Herzog HM, James MG, Cuadra GA. Effects of Oral Commensal Streptococci on Porphyromonas gingivalis Invasion into Oral Epithelial Cells. Dentistry Journal. 2020; 8(2):39. https://0-doi-org.brum.beds.ac.uk/10.3390/dj8020039
Chicago/Turabian StyleHanel, Alyssa N., Hannah M. Herzog, Michelle G. James, and Giancarlo A. Cuadra. 2020. "Effects of Oral Commensal Streptococci on Porphyromonas gingivalis Invasion into Oral Epithelial Cells" Dentistry Journal 8, no. 2: 39. https://0-doi-org.brum.beds.ac.uk/10.3390/dj8020039