Detection of Enteric Viruses on Strawberries and Raspberries Using Capture by Apolipoprotein H
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus Production
2.2. Sequencing of HuNoVs
2.3. Viral Capture Using ApoH
Virus Test Suspensions
2.4. Artificial Contamination of Fruits
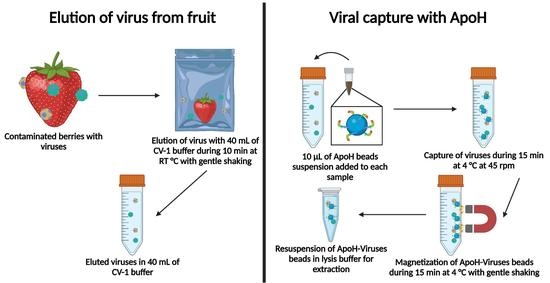
2.5. Elution of Virus from Fruit
2.6. Viral Capture
2.7. Viral RNA Extraction
2.8. RT-qPCR
2.9. Limit of Detection
2.10. Comparison of the ISO and ApoH Methods
2.11. Statistical Analysis
3. Results
3.1. Investigation of the Affinity between Hepatitis A Virus and Apolipoprotein H
3.2. The Limit of Detection of Hepatitis A Virus on Berries
3.3. The Limit of Detection of Norovirus GII.4 on Berries
3.4. Comparison of the ISO and ApoH Methods Using Strawberries
3.5. Comparison of the ISO and ApoH Methods Using Raspberries
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases—Foodborne Disease Burdent Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015; p. 254. [Google Scholar]
- Bozkurt, H.; Phan-Thien, K.Y.; van Ogtrop, F.; Bell, T.; McConchie, R. Outbreaks, occurrence, and control of norovirus and hepatitis a virus contamination in berries: A review. Crit. Rev. Food Sci. Nutr. 2021, 61, 116–138. [Google Scholar] [CrossRef]
- World Health Organization. Global Health Sector Strategy on Viral Hepatitis 2016–2021. Towards Ending Viral Hepatitis; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- World Health Organization; Food and Agriculture Organization of the United Nations. Microbiological Hazards in Fresh Fruits and Vegetables; World Health Organization: Geneva, Switzerland, 2008. [Google Scholar]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization. ISO 15216-1. Microbiology of the Food Chain—Horizontal Method for Determination of Hepatitis A Virus and Norovirus Using Real-Time RT-PCR—Part 1: Method for Quantification; ISO: London, UK, 2017; Available online: https://www.iso.org/standard/65681.html (accessed on 16 December 2021).
- Morton, V.; Jean, J.; Farber, J.; Mattison, K. Detection of noroviruses in ready-to-eat foods by using carbohydrate-coated magnetic beads. Appl. Environ. Microbiol. 2009, 75, 4641–4643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, D.; Zou, S.; Liao, N.; Shi, X.; Chen, J.; Zhang, Y.; Sun, L.; Zhang, R. Evaluation of an Extraction Method for the Detection of GI and GII Noroviruses in Fruit and Vegetable Salads. J. Food Sci. 2018, 83, 393–400. [Google Scholar] [CrossRef]
- Bartsch, C.; Szabo, K.; Dinh-Thanh, M.; Schrader, C.; Trojnar, E.; Johne, R. Comparison and optimization of detection methods for noroviruses in frozen strawberries containing different amounts of RT-PCR inhibitors. Food Microbiol. 2016, 60, 124–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams-Woods, J.; Hartman, G.; Burkhardt, W., III. Bacteriological Analytical Manual Chapter 26B: Detection of Hepatitis A Virus in Foods. BAM 26B; 2017. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-26b-detection-hepatitis-virus-foods (accessed on 16 December 2021).
- Summa, M.; von Bonsdorff, C.H.; Maunula, L. Evaluation of four virus recovery methods for detecting noroviruses on fresh lettuce, sliced ham, and frozen raspberries. J. Virol. Methods 2012, 183, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Grossi, C.; Artusi, C.; Meroni, P.; Borghi, M.O.; Neglia, L.; Lonati, P.A.; Oggioni, M.; Tedesco, F.; De Simoni, M.G.; Fumagalli, S. Beta2 glycoprotein I participates in phagocytosis of apoptotic neurons and in vascular injury in experimental brain stroke. J. Cereb. Blood Flow Metab. 2021, 41, 271678X20984551. [Google Scholar] [CrossRef]
- Liu, Y.; Maiers, J.L.; Rui, Y.; Jiang, X.; Guleng, B.; Ren, J. Apolipoprotein H drives hepatitis B surface antigen retention and endoplasmic reticulum stress during hepatitis B virus infection. Int. J. Biochem. Cell Biol. 2021, 131, 105906. [Google Scholar] [CrossRef]
- Stefas, I.; Tigrett, S.; Dubois, G.; Kaiser, M.; Lucarz, E.; Gobby, D.; Bray, D.; Ellerbrok, H.; Zarski, J.P.; Veas, F. Interactions between Hepatitis C Virus and the Human Apolipoprotein H Acute Phase Protein: A Tool for a Sensitive Detection of the Virus. PLoS ONE 2015, 10, e0140900. [Google Scholar] [CrossRef] [Green Version]
- Almand, E.A.; Goulter, R.M.; Jaykus, L.A. Capture and concentration of viral and bacterial foodborne pathogens using apolipoprotein H. J. Microbiol. Methods 2016, 128, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Mbithi, J.N.; Springthorpe, V.S.; Sattar, S.A. Effect of relative humidity and air temperature on survival of hepatitis A virus on environmental surfaces. Appl. Environ. Microbiol. 1991, 57, 1394–1399. [Google Scholar] [CrossRef] [Green Version]
- Mbithi, J.N.; Springthorpe, V.S.; Boulet, J.R.; Sattar, S.A. Survival of hepatitis A virus on human hands and its transfer on contact with animate and inanimate surfaces. J. Clin. Microbiol. 1992, 30, 757–763. [Google Scholar] [CrossRef] [Green Version]
- Jean, J.; Morales-Rayas, R.; Anoman, M.N.; Lamhoujeb, S. Inactivation of hepatitis A virus and norovirus surrogate in suspension and on food-contact surfaces using pulsed UV light (pulsed light inactivation of food-borne viruses). Food Microbiol. 2011, 28, 568–572. [Google Scholar] [CrossRef]
- Schwarzenbacher, R.; Zeth, K.; Diederichs, K.; Gries, A.; Kostner, G.M.; Laggner, P.; Prassl, R. Crystal structure of human beta2-glycoprotein I: Implications for phospholipid binding and the antiphospholipid syndrome. EMBO J. 1999, 18, 6228–6239. [Google Scholar] [CrossRef]
- Bouma, B.; de Groot, P.G.; van den Elsen, J.M.; Ravelli, R.B.; Schouten, A.; Simmelink, M.J.; Derksen, R.H.; Kroon, J.; Gros, P. Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure. EMBO J. 1999, 18, 5166–5174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolyada, A.; Lee, C.J.; De Biasio, A.; Beglova, N. A novel dimeric inhibitor targeting Beta2GPI in Beta2GPI/antibody complexes implicated in antiphospholipid syndrome. PLoS ONE 2010, 5, e15345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefas, E.; Rucheton, M.; Graafland, H.; Moynier, M.; Sompeyrac, C.; Bahraoui, E.M.; Veas, F. Human plasmatic apolipoprotein H binds human immunodeficiency virus type 1 and type 2 proteins. AIDS Res. Hum. Retrovir. 1997, 13, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Luteijn, R.D.; Praest, P.; Thiele, F.; Sadasivam, S.M.; Singethan, K.; Drijfhout, J.W.; Bach, C.; de Boer, S.M.; Lebbink, R.J.; Tao, S.; et al. A Broad-Spectrum Antiviral Peptide Blocks Infection of Viruses by Binding to Phosphatidylserine in the Viral Envelope. Cells 2020, 9, 1989. [Google Scholar] [CrossRef]
- Goodridge, L.; Goodridge, C.; Wu, J.; Griffiths, M.; Pawliszyn, J. Isoelectric point determination of norovirus virus-like particles by capillary isoelectric focusing with whole column imaging detection. Anal. Chem. 2004, 76, 48–52. [Google Scholar] [CrossRef]
- Kusov, Y.; Gauss-Muller, V.; Morace, G. Immunogenic epitopes on the surface of the hepatitis A virus capsid: Impact of secondary structure and/or isoelectric point on chimeric virus assembly. Virus Res. 2007, 130, 296–302. [Google Scholar] [CrossRef]
- Michen, B.; Graule, T. Isoelectric points of viruses. J. Appl. Microbiol. 2010, 109, 388–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samandoulgou, I.; Fliss, I.; Jean, J. Zeta Potential and Aggregation of Virus-Like Particle of Human Norovirus and Feline Calicivirus Under Different Physicochemical Conditions. Food Environ. Virol. 2015, 7, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Meng, Y.; Hu, C.; Dong, G.; Qu, Y.; Deng, H.; Guo, Y. Mathematical model of Ca2+ concentration, pH, pectin concentration and soluble solids (sucrose) on the gelation of low methoxyl pectin. Food Hydrocoll. 2017, 66, 37–48. [Google Scholar] [CrossRef]


| Sample Volume | Genome Copies/Sample | |||
|---|---|---|---|---|
| 105 | 104 | 103 | 102 | |
| Number of Positive Samples/Number of Samples Tested | ||||
| 1 mL | 4/4 | 4/4 | 3/4 | 1/4 |
| 40 mL | 4/4 | 4/4 | 3/4 | 1/4 |
| Food | Genome Copies/Sample | |||
|---|---|---|---|---|
| 105 | 104 | 103 | 102 | |
| Number of Positive Samples/Number of Samples Tested | ||||
| Fresh Strawberry | 4/4 | 4/4 | 0/4 | 0/4 |
| Frozen Strawberry | 4/4 | 4/4 | 0/4 | 1/4 |
| Fresh Raspberry | 3/4 | 2/4 | 2/4 | 1/4 |
| Frozen Raspberry | 1/4 | 1/4 | 0/4 | 0/4 |
| Food | Genome Copies/Sample | |||
|---|---|---|---|---|
| 105 | 104 | 103 | 102 | |
| Number of Positive Samples/Number of Samples Tested | ||||
| Fresh Strawberry | 4/4 | 0/4 | 1/4 | 0/4 |
| Frozen Strawberry | 4/4 | 1/4 | 2/4 | 0/4 |
| Fresh Raspberry | 2/4 | 2/4 | 0/4 | 1/4 |
| Frozen Raspberry | 1/4 | 0/4 | 2/4 | 0/4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lévesque, A.; Jubinville, E.; Hamon, F.; Jean, J. Detection of Enteric Viruses on Strawberries and Raspberries Using Capture by Apolipoprotein H. Foods 2021, 10, 3139. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123139
Lévesque A, Jubinville E, Hamon F, Jean J. Detection of Enteric Viruses on Strawberries and Raspberries Using Capture by Apolipoprotein H. Foods. 2021; 10(12):3139. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123139
Chicago/Turabian StyleLévesque, Anthony, Eric Jubinville, Fabienne Hamon, and Julie Jean. 2021. "Detection of Enteric Viruses on Strawberries and Raspberries Using Capture by Apolipoprotein H" Foods 10, no. 12: 3139. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10123139