Polar Lipids of Commercial Ulva spp. of Different Origins: Profiling and Relevance for Seaweed Valorization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Proximate Composition Analysis
2.3. Extraction of Polar Lipids
2.4. GC–MS Analysis of Esterified Fatty Acids
2.5. Quantification of Glycolipids and Phospholipids
2.6. Analysis of Polar Lipids by HILIC–MS and MS/MS
2.7. Statistical Analysis
3. Results
3.1. Proximate Composition of Ulva spp. Biomass
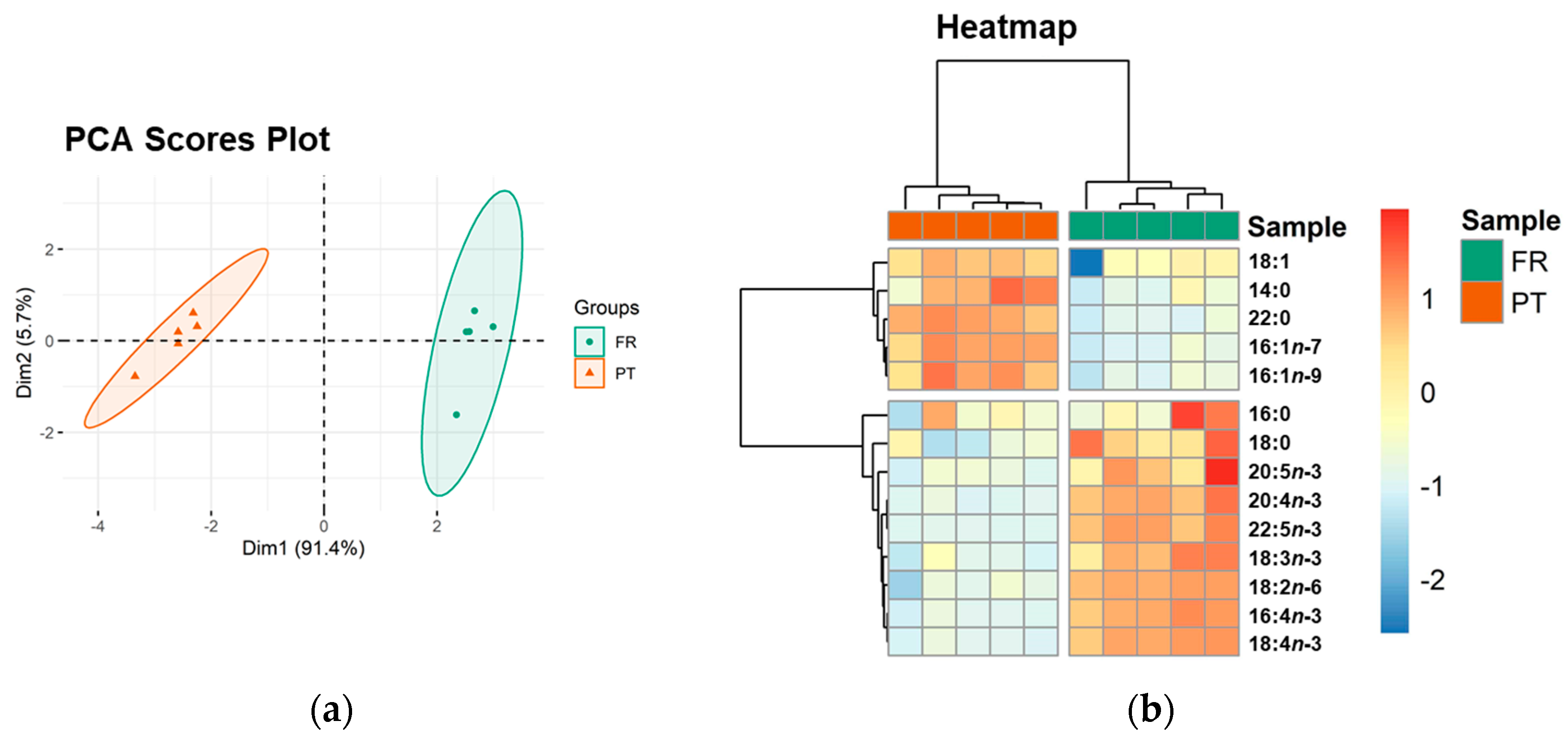
3.2. Esterified Fatty Acid Profile
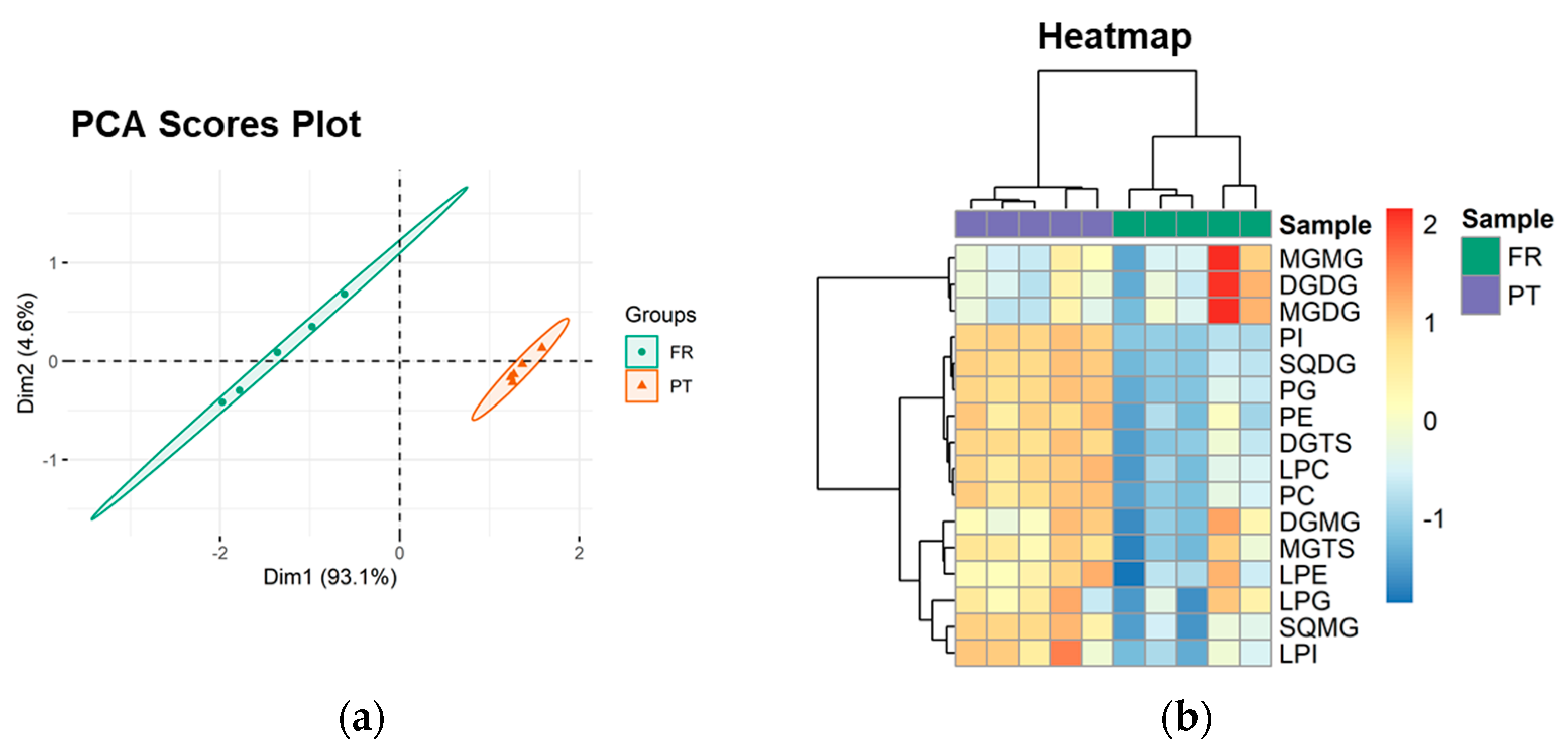
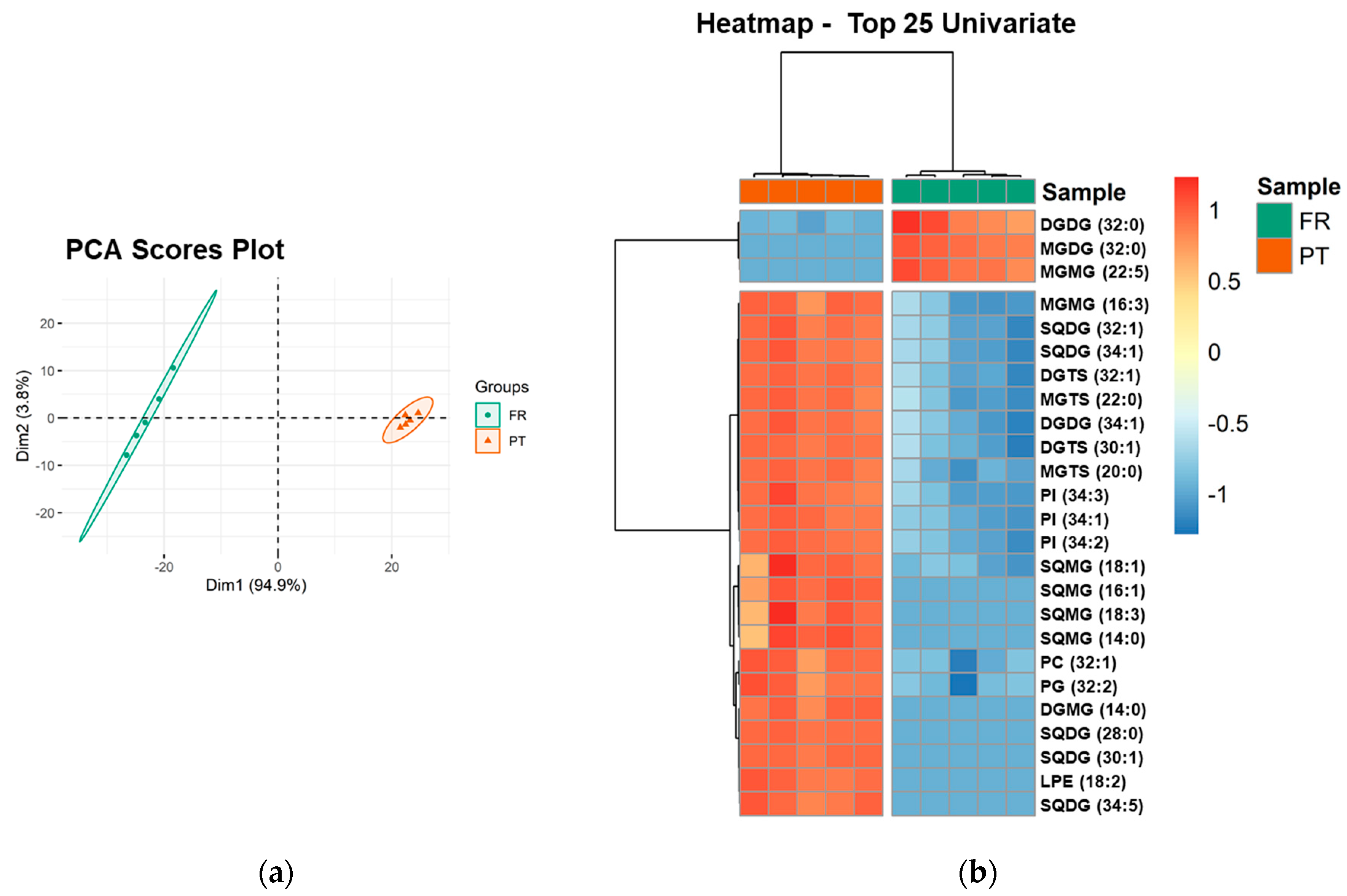
3.3. Polar Lipidome Profile
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- FAO—Food and Agricultural Organization of the United Nations; WHO—World Health Organization. Sustainable Healthy Diets—Guiding Principles; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Tanna, B.; Mishra, A. Metabolites unravel nutraceutical potential of edible seaweeds: An emerging source of functional food. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1613–1624. [Google Scholar] [CrossRef] [Green Version]
- Cherry, P.; O’Hara, C.; Magee, P.J.; McSorley, E.M.; Allsopp, P.J. Risks and benefits of consuming edible seaweeds. Nutr. Rev. 2019, 77, 307–329. [Google Scholar] [CrossRef] [Green Version]
- Stengel, D.B.; Connan, S.; Popper, Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011, 29, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Thelen, J.J.; Fedosejevs, E.; Harwood, J.L. The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 2019, 74, 31–68. [Google Scholar] [CrossRef] [PubMed]
- Eismann, A.I.; Perpetuo Reis, R.; Ferreira da Silva, A.; Negrão Cavalcanti, D. Ulva spp. carotenoids: Responses to environmental conditions. Algal Res. 2020, 48, 101916. [Google Scholar] [CrossRef]
- Guschina, I.A.; Harwood, J.L. Algal lipids and effect of the environment on their biochemistry. In Lipids in Aquatic Ecosystems; Kainz, M., Brett, M.T., Arts, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 1–24. [Google Scholar]
- Gosch, B.J.; Magnusson, M.; Paul, N.A.; de Nys, R. Total lipid and fatty acid composition of seaweeds for the selection of species for oil-based biofuel and bioproducts. GCB Bioenergy 2012, 4, 919–930. [Google Scholar] [CrossRef] [Green Version]
- Swanson, D.; Block, R.; Mousa, S.A. Omega-3 fatty acids EPA and DHA: Health benefits throughout life. Adv. Nutr. 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, A.; Saotome, M.; Iguchi, K.; Maekawa, Y. Marine-derived omega-3 polyunsaturated fatty acids and heart failure: Current understanding for basic to clinical relevance. Int. J. Mol. Sci. 2019, 20, 4025. [Google Scholar] [CrossRef] [Green Version]
- Da Costa, E.; Silva, J.; Mendonça, S.H.; Abreu, M.H.; Domingues, M.R. Lipidomic approaches towards deciphering glycolipids from microalgae as a reservoir of bioactive lipids. Mar. Drugs 2016, 14, 101. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Li, Y.-L.; Shen, W.-Z.; Rui, W.; Ma, X.-J.; Cen, Y.-Z. Antiviral activity of a sulfoquinovosyldiacylglycerol (SQDG) compound isolated from the green alga Caulerpa racemosa. Bot. Mar. 2007, 50, 185–190. [Google Scholar] [CrossRef]
- Arunkumar, K.; Selvapalam, N.; Rengasamy, R. The antibacterial compound sulphoglycerolipid 1-0 palmitoyl-3-0(6′-sulpho-α-quinovopyranosyl)-glycerol from Sargassum wightii Greville (Phaeophyceae). Bot. Mar. 2005, 48, 441–445. [Google Scholar] [CrossRef] [Green Version]
- Banskota, A.H.; Gallant, P.; Stefanova, R.; Melanson, R.; O’Leary, S.J. Monogalactosyldiacylglycerols, potent nitric oxide inhibitors from the marine microalga Tetraselmis chui. Nat. Prod. Res. 2013, 27, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.; Stefanova, R.; Gallant, P.; Osborne, J.A.; Melanson, R.; O’Leary, S.J. Nitric oxide inhibitory activity of monogalactosylmonoacylglycerols from a freshwater microalgae Chlorella sorokiniana. Nat. Prod. Res. 2013, 27, 1028–1031. [Google Scholar] [CrossRef] [PubMed]
- Kendel, M.; Wielgosz-Collin, G.; Bertrand, S.; Roussakis, C.; Bourgougnon, N.; Bedoux, G. Lipid composition, fatty acids and sterols in the seaweeds Ulva armoricana, and Solieria chordalis from Brittany (France): An analysis from nutritional, chemotaxonomic, and antiproliferative activity perspectives. Mar. Drugs 2015, 13, 5606–5628. [Google Scholar] [CrossRef] [PubMed]
- Plouguerné, E.; da Gama, B.A.P.; Pereira, R.C.; Barreto-Bergter, E. Glycolipids from seaweeds and their potential biotechnological applications. Front. Cell Infect. Microbiol. 2014, 4, 174. [Google Scholar] [CrossRef] [PubMed]
- Cotas, J.; Leandro, A.; Pacheco, D.; Gonçalves, A.M.M.; Pereira, L. A comprehensive review of the nutraceutical and therapeutic applications of red seaweeds (Rhodophyta). Life 2020, 10, 19. [Google Scholar] [CrossRef] [Green Version]
- Gullón, B.; Gagaoua, M.; Barba, F.J.; Gullón, P.; Zhang, W.; Lorenzo, J.M. Seaweeds as promising resource of bioactive compounds: Overview of novel extraction strategies and design of tailored meat products. Trends Food Sci. Technol. 2020, 100, 1–18. [Google Scholar] [CrossRef]
- Thiyagarasaiyar, K.; Goh, B.-H.; Jeon, Y.-J.; Yow, Y.-Y. Algae metabolites in cosmeceutical: An overview of current applications and challenges. Mar. Drugs 2020, 18, 323. [Google Scholar] [CrossRef] [PubMed]
- Ścieszka, S.; Klewicka, E. Algae in food: A general review. Crit. Rev. Food Sci. Nutr. 2019, 59, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.S.; Allsopp, P.J.; Magee, P.J.; Gill, C.I.; Nitecki, S.; Strain, C.R.; McSorley, E.M. Seaweed and human health. Nutr. Rev. 2014, 72, 205–216. [Google Scholar] [CrossRef]
- FAO—Food and Agricultural Organization of the United Nations. The Global Status of Seaweed Production, Trade and Utilization; FAO: Rome, Italy, 2018; p. 120. [Google Scholar]
- Mantri, V.A.; Kazi, M.A.; Balar, N.B.; Gupta, V.; Gajaria, T. Concise review of green algal genus Ulva Linnaeus. J. Appl. Phycol. 2020, 32, 2725–2741. [Google Scholar] [CrossRef]
- Bolton, J.J.; Cyrus, M.D.; Brand, M.J.; Joubert, M.; Macey, B.M. Why grow Ulva? Its potential role in the future of aquaculture. Perspect. Phycol. 2016, 3, 113–120. [Google Scholar] [CrossRef]
- Hafting, J.T.; Critchley, A.T.; Cornish, M.L.; Hubley, S.A.; Archibald, A.F. On-land cultivation of functional seaweed products for human usage. J. Appl. Phycol. 2012, 24, 385–392. [Google Scholar] [CrossRef]
- Gadberry, B.A.; Colt, J.; Maynard, D.; Boratyn, D.C.; Webb, K.; Johnson, R.B.; Saunders, G.W.; Boyer, R.H. Intensive land-based production of red and green macroalgae for human consumption in the Pacific Northwest: An evaluation of seasonal growth, yield, nutritional composition, and contaminant levels. Algae 2018, 33, 109–125. [Google Scholar] [CrossRef] [Green Version]
- Mohy El-Din, S.M. Temporal variation in chemical composition of Ulva lactuca and Corallina mediterranea. Int. J. Environ. Sci. Technol. 2019, 16, 5783–5796. [Google Scholar] [CrossRef]
- Moreira, A.S.P.; da Costa, E.; Melo, T.; Sulpice, R.; Cardoso, S.M.; Pitarma, B.; Pereira, R.; Abreu, M.H.; Domingues, P.; Calado, R.; et al. Seasonal plasticity of the polar lipidome of Ulva rigida cultivated in a sustainable integrated multi-trophic aquaculture. Algal Res. 2020, 49, 101958. [Google Scholar] [CrossRef]
- Nelson, M.M.; Phleger, C.F.; Nichols, P.D. Seasonal lipid composition in macroalgae of the northeastern pacific ocean. Bot. Mar. 2002, 45, 58–65. [Google Scholar] [CrossRef]
- Sanina, N.M.; Goncharova, S.N.; Kostetsky, E.Y. Seasonal changes of fatty acid composition and thermotropic behavior of polar lipids from marine macrophytes. Phytochemistry 2008, 69, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Serviere-Zaragoza, E.; Hurtado, M.A.; Manzano-Sarabia, M.; Mazariegos-Villarreal, A.; Reza, M.; Arjona, O.; Palacios, E. Seasonal and interannual variation of fatty acids in macrophytes from the Pacific coast of Baja California Peninsula (Mexico). J. Appl. Phycol. 2015, 27, 1297–1306. [Google Scholar] [CrossRef]
- Da Costa, E.; Ricardo, F.; Melo, T.; Mamede, R.; Abreu, M.H.; Domingues, P.; Domingues, M.R.; Calado, R. Site-specific lipidomic signatures of sea lettuce (Ulva spp., Chlorophyta) hold the potential to trace their geographic origin. Biomolecules 2020, 10, 489. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, C.; Ripol, A.; Afonso, C.; Freire, M.; Varela, J.; Quental-Ferreira, H.; Pousão-Ferreira, P.; Bandarra, N. Fatty acid profiles of the main lipid classes of green seaweeds from fish pond aquaculture. Food Sci. Nutr. 2017, 5, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Alsufyani, T.; Califano, G.; Deicke, M.; Grueneberg, J.; Weiss, A.; Engelen, A.H.; Kwantes, M.; Mohr, J.F.; Ulrich, J.F.; Wichard, T. Macroalgal–bacterial interactions: Identification and role of thallusin in morphogenesis of the seaweed Ulva (Chlorophyta). J. Exp. Bot. 2020, 71, 3340–3349. [Google Scholar] [CrossRef]
- Hoxmark, R.C. Protein composition of different stages in the life cycle of Ulva mutabilis, Føyn. Planta 1976, 130, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Msuya, F.E.; Neori, A. Effect of water aeration and nutrient load level on biomass yield, N uptake and protein content of the seaweed Ulva lactuca cultured in seawater tanks. J. Appl. Phycol. 2008, 20, 1021–1031. [Google Scholar] [CrossRef]
- Silva, A.F.R.; Abreu, H.; Silva, A.M.S.; Cardoso, S.M. Effect of oven-drying on the recovery of valuable compounds from Ulva rigida, Gracilaria sp. and Fucus vesiculosus. Mar. Drugs 2019, 17, 90. [Google Scholar] [CrossRef] [Green Version]
- Lopes, D.; Moreira, A.S.P.; Rey, F.; da Costa, E.; Melo, T.; Maciel, E.; Rego, A.; Abreu, M.H.; Domingues, P.; Calado, R.; et al. Lipidomic signature of the green macroalgae Ulva rigida farmed in a sustainable integrated multi-trophic aquaculture. J. Appl. Phycol. 2019, 31, 1369–1381. [Google Scholar] [CrossRef] [Green Version]
- Angell, A.R.; Mata, L.; de Nys, R.; Paul, N.A. The protein content of seaweeds: A universal nitrogen-to-protein conversion factor of five. J. Appl. Phycol. 2016, 28, 511–524. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Phys. 1959, 37, 911–917. [Google Scholar] [CrossRef] [Green Version]
- Aued-Pimentel, S.; Lago, J.H.G.; Chaves, M.H.; Kumagai, E.E. Evaluation of a methylation procedure to determine cyclopropenoids fatty acids from Sterculia striata St. Hil. Et Nauds seed oil. J. Chromatogr. A 2004, 1054, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Christie, W.W. The Lipid Web. Available online: https://www.lipidmaps.org/resources/lipidweb/index.php?page=ms/methesters/me-arch/index.htm (accessed on 12 November 2020).
- Da Costa, E.; Melo, T.; Moreira, A.S.P.; Bernardo, C.; Helguero, L.; Ferreira, I.; Cruz, M.T.; Rego, A.M.; Domingues, P.; Calado, R.; et al. Valorization of lipids from Gracilaria sp. through lipidomics and decoding of antiproliferative and anti-inflammatory activity. Mar. Drugs 2017, 15, 62. [Google Scholar] [CrossRef] [Green Version]
- Bell, B.M.; Daniels, D.G.H.; Fearn, T.; Stewart, B.A. Lipid compositions, baking qualities and other characteristics of wheat varieties grown in the U.K. J. Cereal Sci. 1987, 5, 277–286. [Google Scholar] [CrossRef]
- Bartlett, E.M.; Lewis, D.H. Spectrophotometric determination of phosphate esters in the presence and absence of orthophosphate. Anal. Biochem. 1970, 36, 159–167. [Google Scholar] [CrossRef]
- Chapman, G.W. A conversion factor to determine phospholipid content in soybean and sunflower crude oils. J. Am. Oil Chem. Soc. 1980, 57, 299–302. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [Green Version]
- Monteiro, J.P.; Rey, F.; Melo, T.; Moreira, A.S.P.; Arbona, J.-F.; Skjermo, J.; Forbord, S.; Funderud, J.; Raposo, D.; Kerrison, P.D.; et al. The unique lipidomic signatures of Saccharina latissima can be used to pinpoint their geographic origin. Biomolecules 2020, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Da Costa, E.; Amaro, H.M.; Melo, T.; Guedes, A.C.; Domingues, M.R. Screening for polar lipids, antioxidant, and anti-inflammatory activities of Gloeothece sp. lipid extracts pursuing new phytochemicals from cyanobacteria. J. Appl. Phycol. 2020, 32, 3015–3030. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 21 January 2020).
- RStudio Team. RStudio: Integrated Development Environment for R. Available online: https://www.rstudio.com/ (accessed on 21 January 2020).
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55, 1–14. [Google Scholar] [CrossRef]
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pcaMethods--A bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Series B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 21 January 2020).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H. ggplot2—Elegant Graphics for Data Analysis. Available online: https://ggplot2.tidyverse.org/ (accessed on 21 January 2020).
- Wickham, H. The split-apply-combine strategy for data analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation. Available online: https://rdrr.io/cran/dplyr/ (accessed on 21 January 2020).
- Wickham, H.; Henry, L. tidyr: Easily Tidy Data with ’Spread()’ and ’Gather()’ Functions. Available online: https://rdrr.io/cran/tidyr/ (accessed on 21 January 2020).
- Slowikowski, K.; Schep, A.; Hughes, S.; Lukauskas, S.; Irisson, J.-O.; Kamvar, Z.N.; Ryan, T.; Christophe, D.; Hiroaki, Y.; Gramme, P. ggrepel: Automatically Position Non-Overlapping Text Labels with ’ggplot2’. Available online: https://rdrr.io/cran/ggrepel/ (accessed on 21 January 2020).
- Fleurence, J.; Gutbier, G.; Mabeau, S.; Leray, C. Fatty acids from 11 marine macroalgae of the French Brittany coast. J. Appl. Phycol. 1994, 6, 527–532. [Google Scholar] [CrossRef]
- Kumari, P.; Kumar, M.; Gupta, V.; Reddy, C.R.K.; Jha, B. Tropical marine macroalgae as potential sources of nutritionally important PUFAs. Food Chem. 2010, 120, 749–757. [Google Scholar] [CrossRef]
- Van Ginneken, V.J.; Helsper, J.P.; de Visser, W.; van Keulen, H.; Brandenburg, W.A. Polyunsaturated fatty acids in various macroalgal species from North Atlantic and tropical seas. Lipids Health Dis. 2011, 10, 10–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaich, H.; Garna, H.; Besbes, S.; Paquot, M.; Blecker, C.; Attia, H. Chemical composition and functional properties of Ulva lactuca seaweed collected in Tunisia. Food Chem. 2011, 128, 895–901. [Google Scholar] [CrossRef]
- Khairy, H.M.; El-Shafay, S.M. Seasonal variations in the biochemical composition of some common seaweed species from the coast of Abu Qir Bay, Alexandria, Egypt. Oceanologia 2013, 55, 435–452. [Google Scholar] [CrossRef] [Green Version]
- Maehre, H.K.; Malde, M.K.; Eilertsen, K.E.; Elvevoll, E.O. Characterization of protein, lipid and mineral contents in common Norwegian seaweeds and evaluation of their potential as food and feed. J. Sci. Food Agric. 2014, 94, 3281–3290. [Google Scholar] [CrossRef] [PubMed]
- Paiva, L.; Lima, E.; Neto, A.I.; Marcone, M.; Baptista, J. Health-promoting ingredients from four selected Azorean macroalgae. Food Res. Int. 2016, 89, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Neto, R.T.; Marçal, C.; Queirós, A.S.; Abreu, H.; Silva, A.M.S.; Cardoso, S.M. Screening of Ulva rigida, Gracilaria sp., Fucus vesiculosus and Saccharina latissima as functional ingredients. Int. J. Mol. Sci. 2018, 19, 2987. [Google Scholar] [CrossRef] [Green Version]
- Roleda, M.Y.; Lage, S.; Aluwini, D.F.; Rebours, C.; Brurberg, M.B.; Nitschke, U.; Gentili, F.G. Chemical profiling of the Arctic sea lettuce Ulva lactuca (Chlorophyta) mass-cultivated on land under controlled conditions for food applications. Food Chem. 2021, 341, 127999. [Google Scholar] [CrossRef]
- Pinheiro, V.F.; Marçal, C.; Abreu, H.; Lopes da Silva, J.A.; Silva, A.M.S.; Cardoso, S.M. Physicochemical changes of air-dried and salt-processed Ulva rigida over storage time. Molecules 2019, 24, 2955. [Google Scholar] [CrossRef] [Green Version]
- Floreto, E.A.T.; Hirata, H.; Ando, S.; Yamasaki, S. Effects of temperature, light intensity, salinity and source of nitrogen on the growth, total lipid and fatty acid composition of Ulva pertusa Kjellman (Chlorophyta). Bot. Mar. 1993, 36, 149–158. [Google Scholar] [CrossRef]
- Abreu, M.H.; Pereira, R.; Yarish, C.; Buschmann, A.H.; Sousa-Pinto, I. IMTA with Gracilaria vermiculophylla: Productivity and nutrient removal performance of the seaweed in a land-based pilot scale system. Aquaculture 2011, 312, 77–87. [Google Scholar] [CrossRef]
- Ulbricht, T.L.V.; Southgate, D.A.T. Coronary heart disease: Seven dietary factors. Lancet 1991, 338, 985–992. [Google Scholar] [CrossRef]
- Dimri, M.; Bommi, P.V.; Sahasrabuddhe, A.A.; Khandekar, J.D.; Dimri, G.P. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis 2010, 31, 489–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, R.D.S.; Campos, M.M. Protective effects of omega-3 fatty acids in cancer-related complications. Nutrients 2019, 11, 945. [Google Scholar] [CrossRef] [Green Version]
- Thompson, G.A. Lipids and membrane function in green algae. Biochim. Biophys. Acta Lipids Lipid Metab. 1996, 1302, 17–45. [Google Scholar] [CrossRef]
- Alsufyani, T.; Engelen, A.H.; Diekmann, O.E.; Kuegler, S.; Wichard, T. Prevalence and mechanism of polyunsaturated aldehydes production in the green tide forming macroalgal genus Ulva (Ulvales, Chlorophyta). Chem. Phys. Lipids 2014, 183, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Lopes, D.; Melo, T.; Rey, F.; Costa, E.; Moreira, A.S.P.; Abreu, M.H.; Domingues, P.; Lillebø, A.I.; Calado, R.; Rosário Domingues, M. Insights of species-specific polar lipidome signatures of seaweeds fostering their valorization in the blue bioeconomy. Algal Res. 2021, 55, 102242. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; Lall, S.P.; Craigie, J.S.; Hafting, J.T.; Critchley, A.T. Polar lipids from the marine macroalga Palmaria palmata inhibit lipopolysaccharide-induced nitric oxide production in RAW264.7 macrophage cells. Phytochemistry 2014, 101, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Banskota, A.H.; Stefanova, R.; Sperker, S.; McGinn, P.J. New diacylglyceryltrimethylhomoserines from the marine microalga Nannochloropsis granulata and their nitric oxide inhibitory activity. J. Appl. Phycol. 2013, 25, 1513–1521. [Google Scholar] [CrossRef]
| FR | PT | Statistical Significance | |
|---|---|---|---|
| Elemental composition | |||
| C (%DW) | 34.12 ± 0.22 | 25.97 ± 0.76 | ** |
| H (%DW) | 6.13 ± 0.21 | 2.82 ± 0.08 | ** |
| N (%DW) | 2.49 ± 0.02 | 1.78 ± 0.05 | ** |
| S (%DW) | 2.09 ± 0.37 | 8.07 ± 0.52 | ** |
| Proximate composition | |||
| Moisture (%) | 8.77 ± 0.25 | 4.45 ± 0.47 | * |
| Ash (%DW) | 13.56 ± 0.52 | 32.50 ± 0.34 | * |
| Proteins (%DW) | |||
| N × 6.25 | 15.59 ± 0.09 | 11.13 ± 0.31 | * |
| N × 5 | 12.47 ± 0.08 | 8.90 ± 0.25 | * |
| Lipids (%DW) | 2.23 ± 0.10 | 1.14 ± 0.12 | ** |
| Carbohydrates and others (%DW) | 68.60 ± 0.56 | 55.23 ± 0.32 | * |
| FR (mg g−1 Ext) | PT (mg g−1 Ext) | Statistical Significance | |
|---|---|---|---|
| Saturated fatty acids (SFA) | 98.37 ± 9.74 | 88.40 ± 5.35 | NS |
| 14:0 | 5.14 ± 0.19 | 6.00 ± 0.41 | * |
| 16:0 | 75.70 ± 8.97 | 69.94 ± 6.03 | NS |
| 18:0 | 13.63 ± 3.08 | 7.05 ± 1.38 | ** |
| 22:0 | 3.90 ± 0.11 | 5.41 ± 0.17 | ** |
| Monounsaturated fatty acids (MUFA) | 27.23 ± 7.03 | 48.89 ± 4.42 | * |
| 16:1n-7 | 3.98 ± 0.33 | 8.29 ± 0.77 | ** |
| 16:1n-9 | 3.75 ± 0.23 | 6.21 ± 0.62 | ** |
| 18:1 | 19.51 ± 6.57 | 34.39 ± 3.08 | ** |
| Polyunsaturated fatty acids (PUFA) | 85.14 ± 9.86 | 27.55 ± 2.07 | * |
| 16:4n-3 | 21.43 ± 3.23 | 5.10 ± 0.44 | ** |
| 18:2n-6 | 8.52 ± 1.08 | 0.91 ± 0.30 | ** |
| 18:3n-3 | 13.29 ± 1.41 | 8.50 ± 0.74 | ** |
| 18:4n-3 | 28.57 ± 3.68 | 6.91 ± 0.58 | ** |
| 20:4n-3 | 2.92 ± 0.21 | 1.75 ± 0.04 | ** |
| 20:5n-3 | 2.66 ±0.24 | 2.17 ± 0.06 | ** |
| 22:5n-3 | 7.75 ± 1.14 | 2.21 ± 0.04 | ** |
| Total PUFA n-3 | 76.62 ± 8.79 | 26.64 ± 1.86 | * |
| PUFA n-6/ PUFA n-3 | 0.11 ± 0.00 | 0.03 ± 0.01 | * |
| Total FA | 210.75 ± 24.07 | 164.84 ± 11.67 | * |
| AI | 0.87 ± 0.10 | 1.23 ± 0.02 | * |
| TI | 0.37 ± 0.04 | 0.60 ± 0.07 | * |
| Polar Lipid Classes | Number of Lipid Species | Major Lipid Species by Class | ||
|---|---|---|---|---|
| FR | PT | FR | PT | |
| Glycolipids | 47 | 53 | ||
| MGMG | 7 | 6 | MGMG (16:0) | MGMG (16:4) |
| DGMG | 4 | 5 | DGMG (16:0) | DGMG (16:0) |
| MGDG | 9 | 8 | MGDG (34:8) | MGDG (34:8) |
| DGDG | 12 | 12 | DGDG (32:0) | DGDG (34:1) |
| SQMG | 2 | 5 | SQMG (16:0) | SQMG (16:0) |
| SQDG | 13 | 17 | SQDG (34:1) | SQDG (34:1) |
| Phospholipids | 49 | 50 | ||
| PC | 12 | 12 | PC (34:1) | PC (36:2) |
| LPC | 9 | 9 | LPC (16:0) | LPC (16:0) |
| PE | 6 | 6 | PE (32:1) | PE (32:1) |
| LPE | 5 | 6 | LPE (16:0) | LPE (16:1) |
| PI | 3 | 3 | PI (34:1) | PI (34:1) |
| LPI | 1 | 1 | LPI (16:0) | LPI (16:0) |
| PG | 9 | 9 | PG (34:4) | PG (34:4) |
| LPG | 4 | 4 | LPG (16:1) | LPG (16:1) |
| Betaine lipids | 51 | 51 | ||
| DGTS | 36 | 36 | DGTS (34:4) | DGTS (32:1) |
| MGTS | 15 | 15 | MGTS (18:3) | MGTS (16:0) |
| Total | 147 | 154 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira, A.S.P.; da Costa, E.; Melo, T.; Lopes, D.; Pais, A.C.S.; Santos, S.A.O.; Pitarma, B.; Mendes, M.; Abreu, M.H.; Collén, P.N.; et al. Polar Lipids of Commercial Ulva spp. of Different Origins: Profiling and Relevance for Seaweed Valorization. Foods 2021, 10, 914. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10050914
Moreira ASP, da Costa E, Melo T, Lopes D, Pais ACS, Santos SAO, Pitarma B, Mendes M, Abreu MH, Collén PN, et al. Polar Lipids of Commercial Ulva spp. of Different Origins: Profiling and Relevance for Seaweed Valorization. Foods. 2021; 10(5):914. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10050914
Chicago/Turabian StyleMoreira, Ana S. P., Elisabete da Costa, Tânia Melo, Diana Lopes, Adriana C. S. Pais, Sónia A. O. Santos, Bárbara Pitarma, Madalena Mendes, Maria H. Abreu, Pi Nyvall Collén, and et al. 2021. "Polar Lipids of Commercial Ulva spp. of Different Origins: Profiling and Relevance for Seaweed Valorization" Foods 10, no. 5: 914. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10050914