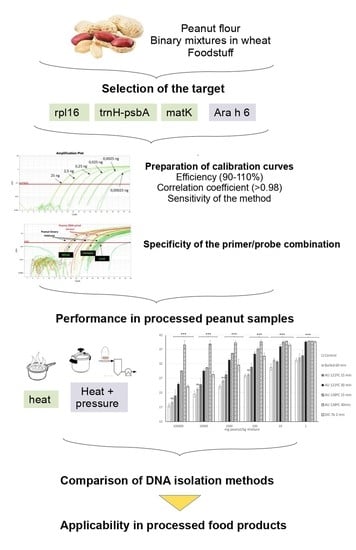
Detection of Peanut Allergen by Real-Time PCR: Looking for a Suitable Detection Marker as Affected by Processing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. Binary Mixtures
2.3. DNA Isolation and Conventional PCR
2.4. Primers, Probes, and Sequencing
2.5. Real-Time PCR
2.6. Statistical Analysis
3. Results and Discussion
3.1. Selection of the Suitable Target
3.2. Effect of Thermal Treatments on Peanut Detection
3.3. Comparative Analysis of DNA Isolation Methods
3.4. Applicability of the Peanut Mat K-Based Detection Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burks, W.; Sampson, H.A.; Bannon, G.A. Peanut allergens. Allergy 1998, 53, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Rabjohn, P.; Helm, E.M.; Stanley, J.S.; West, C.M.; Sampson, H.A.; Burks, A.W.; Bannon, G.A. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J. Clin. Investig. 1999, 103, 535–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maleki, S.J. Food processing: Effects on allergenicity. Curr. Opin. Allergy Clin. Immunol. 2004, 4, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Beyer, K.; Morrow, E.; Li, X.M.; Bardina, L.; Bannon, G.A.; Burks, A.W.; Sampson, H.A. Effects of cooking methods on peanut allergenicity. J. Allergy Clin. Immunol. 2001, 107, 1077–1081. [Google Scholar] [CrossRef]
- Cabanillas, B.; Maleki, S.J.; Rodríguez, J.; Burbano, C.; Muzquiz, M.; Jiménez, M.A.; Pedrosa, M.M.; Cuadrado, C.; Crespo, J.F. Heat and pressure treatments effects on peanut allergenicity. Food Chem. 2012, 132, 360–366. [Google Scholar] [CrossRef]
- Cuadrado, C.; Cabanillas, B.; Pedrosa, M.M.; Muzquiz, M.; Haddad, J.; Allaf, K.; Rodriguez, J.; Crespo, J.F.; Burbano, C. Effect of instant controlled pressure drop on IgE antibody reactivity to peanut, lentil, chickpea and soybean proteins. Int. Arch. Allergy Immunol. 2011, 156, 397–404. [Google Scholar] [CrossRef]
- Pádua, I.; Moreira, A.; Moreira, P.; Melo de Vasconcelos, F.; Barros, R. Impact of the regulation (EU) 1169/2011: Allergen-related recalls in the rapid alert system for food and feed (RASFF) portal. Food Control 2019, 98, 389–398. [Google Scholar] [CrossRef]
- Linacero, R.; Sanchiz, A.; Ballesteros, I.; Cuadrado, C. Application of real-time PCR for tree nut allergen detection in processed foods. Crit. Rev. Food Sci. Nutr. 2020, 60, 1077–1093. [Google Scholar] [CrossRef]
- Chassaigne, H.; Nørgaard, J.V.; Van Hengel, A.J. Proteomics-based approach to detect and identify major allergens in processed peanuts by capillary LC-Q-TOF (MS/MS). J. Agric. Food Chem. 2007, 55, 4461–4473. [Google Scholar] [CrossRef]
- Pafundo, S.; Gullì, M.; Marmiroli, N. Multiplex Real Time PCR using SYBR®GreenERTM for the detection of DNA allergens in food. Anal. Bioanal. Chem. 2010, 396, 1831–1839. [Google Scholar] [CrossRef]
- Brežná, B.; Piknova, L. Real Time PCR Methods for Identification of Animal or Plant Species. Curr. Issues Mol. Biol. 2013, 15, 253–272. [Google Scholar]
- Hird, H.; Lloyd, J.; Goodier, R.; Brown, J.; Reece, P. Detection of peanut using Real Time polymerase chain reaction. Eur. Food Res. Technol. 2003, 217, 265–268. [Google Scholar] [CrossRef]
- Ruiz-Valdepeñas, V.; Pellicanò, A.; Campuzano, S.; Torrente-Rodríguez, R.M.; Reviejo, Á.J.; Cosio, M.S.; Pingarrón, J.M. Electrochemical detection of peanuts at trace levels in foods using a magnetoimmunosensor for the allergenic protein Ara h 2. Sens. Actuators B Chem. 2016, 236, 825–833. [Google Scholar] [CrossRef]
- López-Calleja, I.M.; de la Cruz, S.; Pegels, N.; González, I.; García, T.; Martín, R. Development of a real time PCR assay for detection of allergenic trace amounts of peanut (Arachis hypogaea) in processed foods. Food Control. 2013, 30, 480–490. [Google Scholar] [CrossRef]
- Scaravelli, E.; Brohée, M.; Marchelli, R.; Hengel, A. Development of three Real Time PCR assays to detect peanut allergen residue in processed food products. Eur. Food Res. Technol. 2008, 227, 857–869. [Google Scholar] [CrossRef]
- Cabanillas, B.; Novak, N. Effects of daily food processing on allergenicity. Crit. Rev. Food Sci. Nutr. 2017, 11, 1–12. [Google Scholar] [CrossRef]
- Sanchiz, A.; Pedrosa, M.M.; Guillamón, E.; Arribas, C.; Cabellos, B.; Linacero, R.; Cuadrado, C. Influence of boiling and autoclave processing on the phenolic content, antioxidant activity and functional properties of pistachio, cashew and chestnut flours. LWT—Food Sci. Technol. 2019, 105, 250–256. [Google Scholar] [CrossRef]
- Mattison, C.P.; Bren-Mattison, Y.; Vant-hull, B.; Vargas, A.M.; Wasserman, R.L.; Grimm, C.C. Heat-induced alterations in cashew allergen solubility and IgE binding. Toxicol. Rep. 2016, 3, 244–251. [Google Scholar] [CrossRef] [Green Version]
- Costa, J.; Oliveira, M.B.P.P.; Mafra, I. Effect of thermal processing on the performance of the novel single-tube nested Real Time PCR for the detection of walnut allergens in sponge cakes. Food Res. Int. 2013, 54, 1722–1729. [Google Scholar] [CrossRef]
- Gryson, N. Effect of food processing on plant DNA degradation and PCR-based GMO analysis: A review. Anal. Bioanal. Chem. 2010, 396, 2003–2022. [Google Scholar] [CrossRef]
- Iniesto, E.; Jimenez, A.; Prieto, N.; Cabanillas, B.; Burbano, C.; Pedrosa, M.M.; Rodriguez, J.; Muzquiz, M.; Crespo, J.F.; Cuadrado, C.; et al. Real Time PCR to detect hazelnut allergen coding sequences in processed foods. Food Chem 2013, 138, 1976–1981. [Google Scholar] [CrossRef]
- Linacero, R.; Ballesteros, I.; Sanchiz, Á.; Prieto, N.; Elisa, I.; Martinez, Y.; Pedrosa, M.; Muzquiz, M.; Cabanillas, B.; Rovira, M.; et al. Detection by Real Time PCR of Walnut Allergen Coding Sequences in Processed Foods. Food Chem. 2016, 202, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Prieto, N.; Iniesto, E.; Burbano, C.; Cabanillas, B.; Pedrosa, M.M.; Rovira, M.; Rodr??guez, J.; Muzquiz, M.; Crespo, J.F.; Cuadrado, C.; et al. Detection of almond allergen coding sequences in processed foods by real time PCR. J. Agric. Food Chem. 2014, 62, 5617–5624. [Google Scholar] [CrossRef] [PubMed]
- Sanchiz, A.; Ballesteros, I.; Martin, A.; Rueda, J.; Pedrosa, M.M.; Dieguez, M.D.C.; Rovira, M.; Cuadrado, C.; Linacero, R. Detection of pistachio allergen coding sequences in food products: A comparison of two real time PCR approaches. Food Control 2017, 75, 262–270. [Google Scholar] [CrossRef]
- Scaravelli, E.; Brohée, M.; Marchelli, R.; Van Hengel, A.J. The effect of heat treatment on the detection of peanut allergens as determined by ELISA and Real Time PCR. Anal. Bioanal. Chem. 2009, 395, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wu, P.; Wu, J.; Ping, J.; Wu, J. Advanced DNA-based methods for the detection of peanut allergens in processed food. TrAC Trends Anal. Chem. 2019, 114, 278–292. [Google Scholar] [CrossRef]
- Brzezinski, J.L. Detection of Cashew Nut DNA in spiked Baked goods using Real Time Polymerase Chain Reaction Method. J. AOAC Int. 2006, 89, 1035–1038. [Google Scholar] [CrossRef] [Green Version]
- Demmel, A.; Hupfer, C.; Hampe, E.I.; Busch, U.; Engel, K.H. Development of a Real Time PCR for the detection of lupine DNA (Lupinus species) in foods. J. Agric. Food Chem. 2008, 56, 4328–4332. [Google Scholar] [CrossRef]
- Puente-lelievre, C.; Eischeid, A.C. Development and Evaluation of a Real Time PCR Multiplex Assay for the Detection of Allergenic Peanut Using Chloroplast DNA Markers. J. Agric. Food Chem. 2018, 66, 8623–8629. [Google Scholar] [CrossRef]
- Ladenburger, E.-M.; Dehmer, M.; Grünberg, R.; Waiblinger, H.-U.; Stoll, D.; Bergemann, J. Highly Sensitive Matrix-Independent Quantification of major food allergens peanut and soy by competitive real-time PCR targeting mitochondrial DNA. J. AOAC Int. 2019, 101, 170–184. [Google Scholar] [CrossRef]
- Kim, J.H.; Hong, J.Y.; Moon, J.-C.; Kwon, K.; Jang, C.S. Development of molecular markers for detecting almond, peanut, pine nut, and walnut in commercial food using quantitative real-time PCR. Appl. Biol. Chem. 2018, 61, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Cuadrado, C.; Sanchiz, A.; Vicente, F.; Ballesteros, I.; Linacero, R. Changes Induced by Pressure Processing on Immunoreactive Proteins of Tree Nuts. Molecules 2020, 25, 11. [Google Scholar] [CrossRef] [Green Version]
- Vicente, F.; Sanchiz, A.; Rodríguez-Pérez, R.; Pedrosa, M.; Quirce, S.; Haddad, J.; Besombes, C.; Linacero, R.; Allaf, K.; Cuadrado, C. Influence of instant controlled pressure drop (DIC) on allergenic potential of tree nuts. Molecules 2020, 25, 1742. [Google Scholar] [CrossRef] [Green Version]
- Costa, J.; Melo, V.S.; Santos, C.G.; Oliveira, M.B.P.P.; Mafra, I. Tracing tree nut allergens in chocolate: A comparison of DNA extraction protocols. Food Chem. 2015, 187, 469–476. [Google Scholar] [CrossRef]
- Sanchiz, A.; Ballesteros, I.; Marqués, E.; Dieguez, M.C.; Rueda, J.; Cuadrado, C.; Linacero, R. Evaluation of locked nucleic acid and TaqMan probes for specific detection of cashew nut in processed food by real time PCR. Food Control 2018, 89, 227–234. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Calleja, I.M.; de La Cruz, S.; Gonzalez, I.; Garcia, T.; Martin, R. Market analysis of food products for detection of allergenic walnut (Juglans regia) and pecan (Carya illinoinensis) by Real Time PCR. Food Chem. 2015, 177, 111–119. [Google Scholar] [CrossRef]
- Lopez-Calleja, I.M.; de la Cruz, S.; González, I.; García, T.; Martín, R. Survey of undeclared allergenic pistachio (Pistacia vera) in commercial foods by hydrolysis probe Real Time PCR. Food Control 2014, 39, 49–55. [Google Scholar] [CrossRef]
- Garino, C.; De Paolis, A.; Coïsson, J.D.; Bianchi, D.M.; Decastelli, L.; Arlorio, M. Sensitive and specific detection of pine nut (Pinus spp.) by Real Time PCR in complex food products. Food Chem. 2016, 194, 980–985. [Google Scholar] [CrossRef]
- Brežná, B.; Dudášová, H.; Kuchta, T. A novel Real Time polymerase chain reaction method for the detection of Brazil nuts in food. J. AOAC Int. 2010, 93, 197–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchiz, A.; Cuadrado, C.; Haddad, J.; Linacero, R. Effect of Instant Controlled Pressure Drop (DIC) treatment on the detection of nut allergens by Real Time PCR. Foods 2020, 9, 729. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz, S.; Lopez-Calleja, I.; Martin, R.; Gonzalez, I.; Alcocer, M.; Garcia, T. Recent advances in the detection of allergens in foods. In Food Allergens: Methods and Protocols, Methods in Molecular Biology; Lin, J., Alcocer, M., Eds.; Springer Science: Heidelberg, Germany, 2017; Volume 1592, pp. 263–295. ISBN 978-1-4939-6923-4. [Google Scholar]
- Lopez-Calleja, I.M.; de la Cruz, S.; Pegels, N.; Gonzalez, I.; Garcia, T.; Martin, R. High resolution TaqMan Real Time PCR approach to detect hazelnut DNA encoding for ITS rDNA in foods. Food Chem. 2013, 141, 1872–1880. [Google Scholar] [CrossRef] [PubMed]
- Villa, C.; Costa, J.; Gondar, C.; Oliveira, M.B.P.P.; Mafra, I. Effect of food matrix and thermal processing on the performance of a normalised quantitative Real Time PCR approach for lupine (Lupinus albus) detection as a potential allergenic food. Food Chem. 2018, 262, 251–259. [Google Scholar] [CrossRef] [PubMed]




| Oligonucleotide | Sequence (5′ → 3′) | Amplicon (bp) | Reference |
|---|---|---|---|
| mat K fw | TGGACTCGCCTCTGGTCAT | 104 | |
| mat K rv | CTGCATATCCGCAAATACCG | [30] * | |
| mat K probe | FAM-CATCCCATTAGTAAGCCCGTTTG-BHQ | ||
| trnH-psbA fw | AGGAGCAATAGAAACTGCGT | 68 | |
| trnH-psbA rv | TTTTTGTCTTAAGGGATACGAGT | [30] | |
| trnH-psbA probe | 6FAM-TGATATTGCTCCTTTACTTTCAAAA-BHQ1 | ||
| rpl16 fw | GCGATGGGAACGACGAAAAC | 69 | [30] |
| rpl16 rv | TTAGTTCGTTCCGCCATCCC | ||
| rpl16 probe | 6FAM-ACCTAAGATTCATTTGACGGGA-BHQ1 | ||
| Ara h 6 fw | AGTGCGATAGGTTGCAGGAC | 107 | |
| Ara h 6 rv | AAATCGCAACGCTGTGGTG | This article | |
| Ara h 6 probe | 6FAM-GCAAATGGTGCAGCAGTTCAAGAG-BHQ1 | ||
| Universal 18S fw | CGCGAGAAGTCCACTAAACC | 64 | [36] |
| Universal 18S rv | CCTACGGAAACCTTGTTACGA |
| Peanut Quantity (mg/kg) | rpl 16 | trnH-psbA | mat K | Ara h 6 |
|---|---|---|---|---|
| 100,000 | 15.54 ± 0.02 | 21.45 ± 0.56 | 17.55 ± 0.17 | 25.97 ± 0.02 |
| 10,000 | 18.98 ± 0.07 | 23.55 ± 0.77 | 21.52 ± 0.30 | 28.71 ± 0.11 |
| 1000 | 22.10 ± 0.22 | 26.51 ± 0.33 | 24.17 ± 0.17 | 31.78 ± 0.22 |
| 100 | 26.55 ± 0.02 | 30.39 ± 0.46 | 27.89 ± 0.14 | 34.72 ± 0.05 |
| 10 | 27.24 ± 0.01 | 34.26 ± 0.36 | 30.77 ± 0.25 | 36.65 ± 0.19 |
| 1 | 29.35 ± 0.02 | 37.70 ± 0.17 | 33.22 ± 0.20 | 37.61 ± 1.35 * |
| 0.5 | - | - | 32.74 ± 0.15 * | - |
| 0.1 | - | - | 33.87 ± 0.58 * | - |
| Slope | −3.04 | −3.35 | −3.14 | −2.74 |
| Efficiency (%) | 113.52 | 98.81 | 108.30 | 131.95 |
| R2 | 0.970 | 0.991 | 0.995 | 0.994 |
| Common Name | Scientific Name | Ct ± SD | |||
|---|---|---|---|---|---|
| rpl16 | trnH-psbA | mat K | Ara h6 | ||
| Apple | Malus domestica | - | - | 39.70 ± 0.42 | - |
| Lentil | Lens culinaris | 28.30 ± 1.00 | N.D. | 39.51 ± 0.68 | - |
| Lemon | Citrus x limon | - | - | 37.24 ± 0.21 | - |
| Lupin | Lupinus albus | 29.10± 0.25 | - | 37.71 ± 1.22 | 34.60 ± 0.28 |
| Almond | Prunus dulcis | - | N.D. | 37.79 ± 1.24 | - |
| Green bean | Phaseolus vulgaris | 38.7 ± 0.14 | N.D. | 38.25 ± 0.79 | - |
| Kiwi | Actinia deliciosa | - | - | N.D. | - |
| Carob | Ceratonia siliqua | 28.54 ± 0.46 | - | 35.46 ± 2.48 | - |
| Walnut | Juglans regia | - | N.D. | 35.55 ± 0.26 | - |
| Pear | Pyrus pyrifolia | - | - | N.D. | - |
| Fababean | Vicia faba | 28.44 ± 0.70 | N.D. | 36.20 ± 1.62 | - |
| Chickpea | Cicer arietinum | 26.83 ± 1.57 | N.D. | 37.73 ± 2.60 | 31.62 ± 0.01 |
| Soy | Glycine max | 29.44 ± 0.03 | N.D. | 39.06 ± 1.11 | - |
| Pistachio | Pistacia vera | - | 35.33 ± 0.94 | 34.44 ± 0.27 | 34.90± 0.08 |
| Hazelnut | Corylus avellana | - | 36.97 ± 0.38 | 37.51 ± 0.48 | 34.22 ± 0.01 |
| Cashew | Anacardium occidentale | - | 37.39 ± 1.48 | 37.39 ± 1.48 | - |
| Grass pea | Lathyrus sativus | 37.08 ± 0.83 | - | 37.77 ± 0.89 | - |
| Chestnut | Castanea sativa | - | - | N.D. | - |
| Pea | Pisum sativum | N.D. | N.D. | 38.47 ± 0.88 | 33.74 ± 0.14 |
| Wheat | Triticum spelta | N.D. | 35.70 ± 0.30 | N.D. | 35.17 ± 0.43 |
| Food | Food Allergen Declaration | Ct ± SD | Peanut mg/kg * |
|---|---|---|---|
| Cereal Bar I | Peanut (35%), Hazelnut (24%) | 17.97 ± 0.26 | >105 |
| Cereal Muesli | Tree nut and peanut traces | 38.73 ± 0.76 | <LOQ |
| Cereal Bar II | Almond and tree nuts | 32.40 ± 0.29 | 2.5 |
| Cereal Bar III | Hazelnut (10%), Almond and Peanut traces | 33.74 ± 0.29 | <1 |
| Cereal Bar IV | May content tree nut traces | 35.90 ± 0.94 | <LOQ |
| Chocolate with pistachio | Pistachio (5%), Almond, Hazelnut, tree nut traces | 36.80 ± 0.43 | <LOQ |
| Vegetal Burger | May content tree nut traces | 39.17 ± 0.96 | <LOQ |
| Sausage with walnut | Walnuts | N.D. | N.D. |
| Chocolate | Almond and Hazelnut traces | 37.99 ± 0.33 | <LOQ |
| Chocolate Bar I | Tree nut traces | 32.42 ± 0.16 | 2.5 |
| Chocolate Bar II | Not declared | 31.32 ± 0.21 | 5.7 |
| Cookies with fiber | Not declared | 31.02 ± 0.30 | 7.1 |
| Cookies with chocolate | Not declared | 32.25 ± 0.46 | 5.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchiz, A.; Sánchez-Enciso, P.; Cuadrado, C.; Linacero, R. Detection of Peanut Allergen by Real-Time PCR: Looking for a Suitable Detection Marker as Affected by Processing. Foods 2021, 10, 1421. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10061421
Sanchiz A, Sánchez-Enciso P, Cuadrado C, Linacero R. Detection of Peanut Allergen by Real-Time PCR: Looking for a Suitable Detection Marker as Affected by Processing. Foods. 2021; 10(6):1421. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10061421
Chicago/Turabian StyleSanchiz, Africa, Paulina Sánchez-Enciso, Carmen Cuadrado, and Rosario Linacero. 2021. "Detection of Peanut Allergen by Real-Time PCR: Looking for a Suitable Detection Marker as Affected by Processing" Foods 10, no. 6: 1421. https://0-doi-org.brum.beds.ac.uk/10.3390/foods10061421