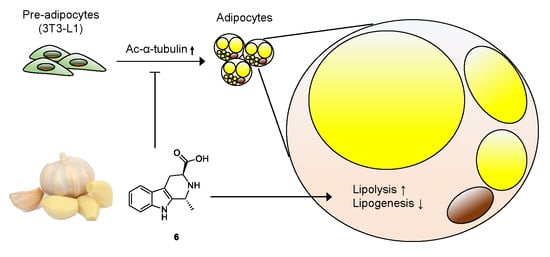
Anti-adipogenic Effect of β-Carboline Alkaloids from Garlic (Allium sativum)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Extraction of Garlic Sample and Isolation of Compounds
2.2. Cell Culture and Differentiation
2.3. Cell Counting
2.4. Immunoblotting
2.5. Reverse Transcription and Quantitative Real-Time PCR (RT-qPCR)
2.6. Oil Red O Staining
2.7. Statistical Analysis
3. Results and Discussion
3.1. Identification of Compounds
3.2. Effects of Compounds 1–6 on Adipogenesis
3.3. Effects of Compound 6 on the Early Stages of Adipogenesis
3.4. Effects of Compound 6 on Lipid Metabolism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Angiosperm Phylogeny Group. An update of the Angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar] [CrossRef] [Green Version]
- Rivlin, R.S. Historical perspective on the use of garlic. J. Nutr. 2001, 132, 951S–954S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iciek, M.; Kwiecieri, I.; Wlodek, L. Biological properties of garlic and garlic-derived organosulfur compounds. Environ. Mol. Mutagen. 2009, 50, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Shamseer, L.; Charrois, T.L.; Vohra, S. Complementary, holistic and integrative medicine: Garlic. Pediatr. Rev. 2006, 27, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Jacob, B.; Narendhirakannan, R.T. Role of medicinal plants in the management of diabetes mellitus: A review. 3 Biotech. 2019, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Pittler, M.H.; Ernst, E. Clinical effectiveness of garlic (Allium sativum). Mol. Nutr. Food Res. 2007, 51, 1382–1385. [Google Scholar] [CrossRef]
- Štajner, D.; Milić, N.; Čanadanović-Brunet, J.; Kapor, A.; Štajner, M.; Popović, B.M. Exploring Allium species as a source of potential medicinal agents. Phytother. Res. 2006, 20, 581–584. [Google Scholar] [CrossRef]
- Lanzotti, V. The analysis of onion and garlic. J. Chromatogr. A 2006, 1112, 3–22. [Google Scholar] [CrossRef] [Green Version]
- Amagase, H.; Petesch, B.L.; Matsuura, H.; Kasuga, S.; Itakura, Y. Intake of garlic and its bioactive components. J. Nutr. 2001, 131, 955S–962S. [Google Scholar] [CrossRef]
- Lawson, L.D.; Ransom, D.K.; Hughes, B.G. Inhibition of whole blood platelet-aggregation by compounds in garlic clove extracts and commercial garlic products. Thromb. Res. 1992, 65, 141–156. [Google Scholar] [CrossRef]
- Hussain, S.P.; Jannu, L.N.; Rao, A.R. Chemopreventive action of garlic on methylcholanthrene-induced carcinogenesis in the uterine cervix of mice. Cancer Lett. 1990, 49, 175–180. [Google Scholar] [CrossRef]
- Pinto, J.T.; Lapsia, S.; Shah, A.; Santiago, H.; Kim, G. Antiproliferative effects of garlic-derived and other allium related compounds. Adv. Exp. Med. Biol. 2001, 492, 83–106. [Google Scholar] [PubMed]
- Key, T.J.; Silcocks, P.B.; Davey, G.K.; Appleby, P.N.; Bishop, D.T. A case-control study of diet and prostate cancer. Br. J. Cancer 1997, 76, 678–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milner, J.A. Garlic: Its anticarcinogenic and antitumorigenic properties. Nutr. Rev. 1996, 54, S82–S86. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, K.C. Therapeutic actions of garlic constituents. Med. Res. Rev. 1996, 16, 111–124. [Google Scholar] [CrossRef]
- Yeh, Y.Y. Garlic phytochemicals in disease prevention and health promotion: An overview. New Drug Clin. 1996, 45, 441–450. [Google Scholar]
- Hirsch, K.; Danilenko, M.; Giat, J.; Miron, T.; Rabinkov, A.; Wilchek, M.; Mirelman, D.; Levy, H.; Sharoni, Y. Effect of purified allicin, the major ingredient of freshly crushed garlic, on cancer cell proliferation. Nutr. Cancer 2000, 38, 245–254. [Google Scholar] [CrossRef]
- Shang, A.; Cao, S.; Xu, X.; Gan, R.; Tang, G.; Corke, H.; Mavumengwana, V.; Li, H. Bioactive compounds and biological functions of garlic (Allium sativum L.). Foods 2019, 8, 246. [Google Scholar] [CrossRef] [Green Version]
- Corzo-Martınez, M.; Corzo, N.; Villamiel, M. Biological properties of onions and garlic. Trends Food Sci. Technol. 2007, 18, 609–625. [Google Scholar] [CrossRef]
- Lanzotti, V.; Barile, E.; Antignani, V.; Bonanomi, G.; Scala, F. Antifungal saponins from bulbs of garlic, Allium sativum L. var. Voghiera. Phytochemistry 2012, 78, 126–134. [Google Scholar] [CrossRef]
- Ichikawa, M.; Yoshida, J.; Ide, N.; Sasaoka, T.; Yamaguchi, H.; Ono, K. Tetrahydro-beta-carboline derivatives in aged garlic extract show antioxidant properties. J. Nutr. 2006, 136, 726S–731S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- So, H.M.; Eom, H.J.; Lee, D.; Kim, S.; Kang, K.S.; Lee, I.K.; Baek, K.-H.; Park, J.Y.; Kim, K.H. Bioactivity evaluations of betulin identified from the bark of Betula platyphylla var. japonica for cancer therapy. Arch. Pharm. Res. 2018, 41, 815–822. [Google Scholar] [PubMed]
- Baek, S.C.; Choi, E.; Eom, H.J.; Jo, M.S.; Kim, S.; So, H.M.; Kim, S.H.; Kang, K.S.; Kim, K.H. LC/MS-based analysis of bioactive compounds from the bark of Betula platyphylla var japonica and their effects on regulation of adipocyte and osteoblast differentiation. Nat. Prod. Sci. 2018, 24, 235–240. [Google Scholar]
- Yu, J.S.; Roh, H.-S.; Baek, K.-H.; Lee, S.; Kim, S.; So, H.M.; Moon, E.; Pang, C.; Jang, T.S.; Kim, K.H. Bioactivity-guided isolation of ginsenosides from Korean Red Ginsengwith cytotoxic activity against human lung adenocarcinoma cells. J. Ginseng Res. 2018, 42, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Ambati, S.; Yang, J.Y.; Rayalam, S.; Park, H.J.; Della-Fera, M.A.; Baile, C.A. Ajoene exerts potent effects in 3T3-L1 adipocytes by inhibiting adipogenesis and inducing apoptosis. Phytother. Res. 2009, 23, 513–518. [Google Scholar] [CrossRef]
- Lii, C.K.; Huang, C.Y.; Chen, H.W.; Chow, M.Y.; Lin, Y.R.; Huang, C.S.; Tsai, C.W. Diallyl trisulfide suppresses the adipogenesis of 3T3-L1 preadipocytes through ERK activation. Food Chem. Toxicol. 2012, 50, 478–484. [Google Scholar] [CrossRef]
- Ban, J.O.; Lee, D.H.; Kim, E.J.; Kang, J.W.; Kim, M.S.; Cho, M.C.; Jeong, H.S.; Kim, J.W.; Yang, Y.; Hong, J.T.; et al. Antiobesity effects of a sulfur compound thiacremonone mediated via down-regulation of serum triglyceride and glucose levels and lipid accumulation in the liver of db/db mice. Phytother. Res. 2012, 26, 1265–1271. [Google Scholar] [CrossRef]
- Kim, E.J.; Lee, D.H.; Kim, H.J.; Lee, S.J.; Ban, J.O.; Cho, M.C.; Jeong, H.S.; Yang, Y.; Hong, J.T.; Yoon, D.Y. Thiacremonone, a sulfur compound isolated from garlic, attenuates lipid accumulation partially mediated via AMPK activation in 3T3-L1 adipocytes. J. Nutr. Biochem. 2012, 23, 1552–1558. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yahara, S.; Kato, K.; Nohara, T. Studies on the constituents of the water soluble portion in Asiasari radix. Shoyakugaku Zasshi 1990, 44, 331–334. [Google Scholar]
- Orihara, Y.; Furuya, T.; Hashimoto, N.; Deguchi, Y.; Tokoro, K.; Kanisawa, T. Biotransformation of isoeugenol and eugenol by cultured cells of Eucalyptus perriniana. Phytochemistry 1992, 31, 827–831. [Google Scholar] [CrossRef]
- Wang, X.; Liu, R.; Yang, Y.; Zhang, M. Isolation, purification and identification of antioxidants in an aqueous aged garlic extract. Food Chem. 2015, 187, 37–43. [Google Scholar] [CrossRef]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef]
- Yang, W.; Guo, X.; Thein, S.; Xu, F.; Sugii, S.; Baas, P.W.; Radda, G.K.; Han, W. Regulation of adipogenesis by cytoskeleton remodelling is facilitated by acetyltransferase MEC-17-dependent acetylation of α-tubulin. Biochem. J. 2013, 449, 605–612. [Google Scholar] [CrossRef] [Green Version]
- Spiegelman, B.M.; Farmer, S.R. Decreases in tubulin and actin gene expression prior to morphological differentiation of 3T3 adipocytes. Cell 1982, 29, 53–60. [Google Scholar] [CrossRef]
- Spiegelman, B.M.; Flier, J.S. Obesity and the Regulation of Energy Balance. Cell 2001, 104, 531–543. [Google Scholar] [CrossRef] [Green Version]
- Smith, U.; Kahn, B.B. Adipose tissue regulates insulin sensitivity: Role of adipogenesis, de novo lipogenesis and novel lipids. J. Intern. Med. 2016, 280, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Piechowska, P.; Zawirska-Wojtasiak, R.; Mildner-Szkudlarz, S. Bioactive β-Carbolines in Food: A Review. Nutrients 2019, 11, 814. [Google Scholar] [CrossRef] [Green Version]
- Ferraz, C.A.A.; de Oliveira Júnior, R.G.; Picot, L.; da Silva Almeida, J.R.G.; Nunes, X.P. Pre-clinical investigations of β-carboline alkaloids as antidepressant agents: A systematic review. Fitoterapia 2019, 137, 104196. [Google Scholar] [CrossRef]
- Zhao, W.Y.; Zhou, W.Y.; Chen, J.J.; Yao, G.D.; Lin, B.; Wang, X.B.; Huang, X.X.; Song, S.J. Enantiomeric β-carboline dimers from Picrasma quassioides and their anti-hepatoma potential. Phytochemistry 2019, 159, 39–45. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, D. Recent Advances of Natural and Synthetic β-Carbolines as Anticancer Agents. Anticancer Agents Med. Chem. 2015, 15, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Forcioli-Conti, N.; Estève, D.; Bouloumié, A.; Dani, C.; Peraldi, P. The size of the primary cilium and acetylated tubulin are modulated during adipocyte differentiation: Analysis of HDAC6 functions in these processes. Biochimie 2016, 124, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Shi, S.; Wang, H.; Liao, K. Growth arrest induces primary-cilium formation and sensitizes IGF-1-receptor signaling during differentiation induction of 3T3-L1 preadipocytes. J. Cell Sci. 2009, 122, 2760–2768. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Gene | Forward | Reverse |
|---|---|---|
| β-Actin | 5′-ACGGCCAGGTCATCACTATTG-3′ | 5′-TGGATGCCACAGGATTCCA-3′ |
| Adipsin | 5‘-CATGCTCGGCCCTACATG-3′ | 5‘-CACAGAGTCGTCATCCGTCAC-3′ |
| Adipoq | 5′-TGTTCCTCTTAATCCTGCCCA-3′ | 5′-CCAACCTGCACAAGTTCCCTT-3′ |
| ATGL | 5′-TTCACCATCCGCTTGTTGGAG-3′ | 5′-AGATGGTCACCCAATTTCCTC-3′ |
| C/EBPα | 5′-CTCCCAGAGGACCAATGAAA-3′ | 5′-AAGTCTTAGCCGGAGGAAGC-3′ |
| C/EBPβ | 5′-GGACAAGCTGAGCGACGAGTA-3′ | 5′-CAGCTGCTCCACCTTCTTCTG-3′ |
| Fabp4 | 5‘-AAGGTGAAGAGCATCATAACCCT-3′ | 5‘-TCACGCCTTTCATAACACATTCC-3′ |
| HSL | 5′-CACAAAGGCTGCTTCTACGG-3′ | 5′-GGAGAGAGTCTGCAGGAACG-3′ |
| PPARγ | 5‘-GCATGGTGCCTTCGCTGA-3′ | 5‘-TGGCATCTCTGTGTCAACCATG-3′ |
| SREBP1 | 5′-AACGTCACTTCCAGCTAGAC-3′ | 5′-CCACTAAGGTGCCTACAGAGC-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, S.C.; Nam, K.H.; Yi, S.A.; Jo, M.S.; Lee, K.H.; Lee, Y.H.; Lee, J.; Kim, K.H. Anti-adipogenic Effect of β-Carboline Alkaloids from Garlic (Allium sativum). Foods 2019, 8, 673. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8120673
Baek SC, Nam KH, Yi SA, Jo MS, Lee KH, Lee YH, Lee J, Kim KH. Anti-adipogenic Effect of β-Carboline Alkaloids from Garlic (Allium sativum). Foods. 2019; 8(12):673. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8120673
Chicago/Turabian StyleBaek, Su Cheol, Ki Hong Nam, Sang Ah Yi, Mun Seok Jo, Kwang Ho Lee, Yong Hoon Lee, Jaecheol Lee, and Ki Hyun Kim. 2019. "Anti-adipogenic Effect of β-Carboline Alkaloids from Garlic (Allium sativum)" Foods 8, no. 12: 673. https://0-doi-org.brum.beds.ac.uk/10.3390/foods8120673