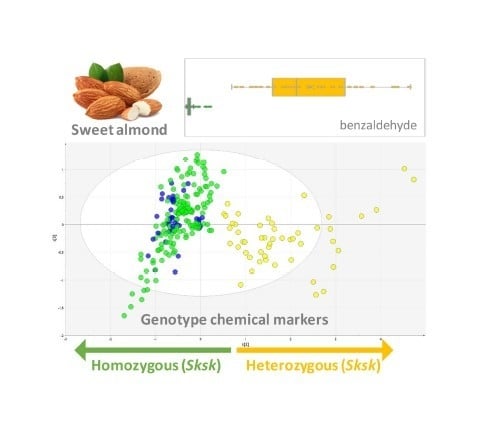
Chemical Markers to Distinguish the Homo- and Heterozygous Bitter Genotype in Sweet Almond Kernels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Chemical Reagents
2.3. Sample Preparation and Solid Phase Microextraction (SPME) Conditions
2.4. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
2.5. Statistical Analysis
3. Results and Discussion
3.1. Optimization of SPME-GC-MS Method for the Assessment of Volatile Compounds
3.2. Univariate Statistical Analysis of Raw Almond Volatile Components
3.3. Multivariate Statistical Analysis of Raw Almond Volatile Components
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT (2018). Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 27 March 2020).
- Socias, R.; Kodad, O.; Alonso, J.M.; Gradziel, T.M. Almond quality: A breeding perspective. Hort. Rev. 2008, 34, 197–238. [Google Scholar]
- Romero, A. Almond Quality Requirements for Industrial Purposes—Its Relevance for the Future Acceptance of New Cultivars from Breeding Programs. Rev. Acta Hort. 2014, 1028, 213–219. [Google Scholar] [CrossRef]
- Batlle, I.; Dicenta, F.; Rafael Socias i Company; Gradziel, T.M.; Wirthensohn, M.; Duval, H.; Vargas, F.J. Classical genetics and breeding. In Almond: Botany, Production and Uses; Rafael Socias i Company, Gradziel, T.M., Eds.; CABI: Oxfordshire, UK, 2017; pp. 111–148. [Google Scholar]
- Gradziel, T.M.; Curtis, R.; Rafael Socias i Company. Production and growing regions. In Almond: Botany, Production and Uses; Rafael Socias i Company, Gradziel, T.M., Eds.; CABI: Oxfordshire, UK, 2017; pp. 70–86. [Google Scholar]
- Wirthensohn, M.; Iannamico, L. Almond in the southern hemisphere. In Almond: Botany, Production and Uses; Rafael Socias i Company, Gradziel, T.M., Eds.; CABI: Oxfordshire, UK, 2017; pp. 87–110. [Google Scholar]
- Sanchez-Perez, R.; Jørgensen, K.; Olsen, C.E.; Dicenta, F.; Lindberg Møller, B. Bitterness in Almonds. Rev. Plant Physiol. 2008, 146, 1040–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, E.E. Cyanogenic compounds. Annu. Rev. Plant. Physiol. 1980, 31, 433–451. [Google Scholar] [CrossRef]
- Dicenta, F.; Martínez-Gómez, P.; Grané, N.; Martín, M.L.; León, A.; Cánovas, J.A.; Berenguer, V. Relationship between cyanogenic compounds in kernels, leaves, and roots of sweet and bitter kernelled almonds. J. Agric. Food Chem. 2002, 50, 2149–2152. [Google Scholar] [CrossRef]
- Wirthensohn, M.G.; Chin, W.L.; Franks, T.K.; Baldock, G.; Ford, C.M.; Sedgley, M. Characterising the flavour phenotypes of almond (Prunus dulcis Mill.) kernels. J. Hort. Sci. Biotech. 2008, 83, 462–468. [Google Scholar] [CrossRef]
- Frehner, M.; Scalet, M.; Conn, E.E. Pattern of the cyanide-potential in developing fruits. Plant Physiol. 1990, 94, 28–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dicenta, F.; García, J.E. Inheritance of kernel flavour in almond. Heredity 1993, 70, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Dicenta, F.; Martínez-Gómez, P.; Ortega, E.; Duval, H. Cultivar pollinizer does not affect almond flavor. Hort. Sci. 2000, 35, 1153–1154. [Google Scholar] [CrossRef] [Green Version]
- Heppner, J. The factor for bitterness in the sweet almond. Genetics 1923, 8, 390–392. [Google Scholar] [PubMed]
- Heppner, J. Further evidence on the factor for bitterness in the sweet almond. Genetics 1926, 11, 605–606. [Google Scholar] [PubMed]
- Dicenta, F.; Ortega, E.; Martinez-Gomez, P. Use of recessive homozygous genotypes to assess the genetic control of kernel bitterness in almond. Euphytica 2007, 153, 221–225. [Google Scholar] [CrossRef]
- Sánchez-Pérez, R.; Howard, W.; Garcia-Mas, J.; Arús, P.; Martínez-Gómez, P.; Dicenta, F. Molecular markers for kernel bitterness in almond. Tree Genet. Genomes 2010, 6, 237–245. [Google Scholar] [CrossRef]
- Sánchez-Pérez, R.; Pavan, S.; Mazzeo, R.; Moldovan, C.; Aiese Cigliano, R.; Del Cueto, J.; Ricciardi, F.; Lotti, C.; Ricciardi, L.; Dicenta, F.; et al. Mutation of a bHLH transcription factor allowed almond domestication. Science 2019, 364, 1095–1098. [Google Scholar] [CrossRef] [PubMed]
- Alioto, T.; Alexiou, K.G.; Bardil, A.; Barteri, F.; Castanera, R.; Cruz, F.; Dhingra, A.; Duval, H.; Fernández i Martí, A.; Frias, L.; et al. Transposons played a major role in the diversification between the closely related almond and peach genomes: Results from the almond genome sequence. Plant J. 2020, 101, 455–472. [Google Scholar] [CrossRef] [Green Version]
- Vargas, F.J.; Romero, M.A.; Batlle, I. Kernel taste inheritance in almond. In XIGREMPA Seminar on Pistachios and Almonds. Cahiers Options Méditerranéennes; Ak, B.E., Ed.; CIHEAM: Zaragoza, Spain, 2001; Volume 56, pp. 129–134. [Google Scholar]
- Ricciardi, F.; Del Cueto, J.; Bardaro, N.; Mazzeo, R.; Ricciardi, L.; Dicenta, F.; Sánchez-Pérez, R.; Pavan, S.; Lotti, C. Synteny-Based Development of CAPS Markers Linked to the Sweet kernel LOCUS, Controlling Amygdalin Accumulation in Almond (Prunus dulcis (Mill.) D.A.Webb). Genes 2020, 9, 385. [Google Scholar] [CrossRef] [Green Version]
- Arrázola, G.; Sánchez-Pérez, R.; Dicenta, F.; Grané, N. Content of the cyanogenic glucoside amygdalin in almond seeds related to the bitterness genotype. Agron. Colomb. 2012, 30, 260–265. [Google Scholar]
- Lee, J.; Zhang, G.; Wood, E.; Rogel Castillo, C.; Mitchell, A.E. Quantification of Amygdalin in Nonbitter, Semibitter, and Bitter Almonds (Prunus dulcis) by UHPLC-(ESI) QqQ MS/MS. J. Agric. Food Chem. 2013, 61, 7754–7759. [Google Scholar] [CrossRef]
- Cortés, V.; Talens, P.; Barat, M.; Lerma-García, M.J. Potential of NIR spectroscopy to predict amygdalin content established by HPLC in intact almonds and classification based on almond bitterness. Food Control 2018, 91, 68–75. [Google Scholar] [CrossRef]
- King, E.S.; Chapman, D.M.; Luo, K.; Ferris, S.; Huang, G.; Mitchell, A.E. Defining the Sensory Profiles of Raw Almond (Prunus dulcis) Varieties and the Contribution of Key Chemical Compounds and Physical Properties. J. Agric. Food Chem. 2019, 67, 3229–3241. [Google Scholar] [CrossRef] [Green Version]
- Kwak, J.; Faranda, A.; Henkin, J.H.; Gallagher, M.; Preti, G.; McGovern, P.E. Volatile organic compounds released by enzymatic reactions in raw nonpareil almond kernel. Eur. Food Res. Technol. 2015, 241, 441–446. [Google Scholar] [CrossRef]
- Oliveira, I.; Malheiro, R.; Meyer, A.S.; Pereira, J.A.; Gonçalves, B. Application of chemometric tools for the comparison of volatile profile from raw and roasted regional and foreign almond cultivars (Prunus dulcis). J. Food Sci. Technol. 2019, 56, 3764–3776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dicenta, F.; Sánchez-Pérez, R.; Rubio, M.; Egea, J.; Batlle, I.; Miarnau, X.; Palasciano, M.; Lipari, E.; Confolent, C.; Martínez-Gómez, P.; et al. The origin of the self-compatible almond ‘Guara’. Sci. Hortic. 2015, 197, 1–4. [Google Scholar] [CrossRef]
- Xiao, L.; Lee, J.; Zhang, G.; Ebeler, S.E.; Wickramasinghe, N.; Seiber, J.; Mitchell, A.E. HS-SPME GC/MS characterization of volatiles in raw and dry-roasted almonds (Prunus dulcis). Food Chem. 2014, 151, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Franklin, L.M.; Mitchell, A.E. Review of the Sensory and Chemical Characteristics of Almond (Prunus dulcis) Flavor. J. Agric. Food Chem. 2019, 67, 2743–2753. [Google Scholar] [CrossRef] [Green Version]
- Luo, K.K.; Kim, D.A.; Mitchell-Silbaugh, K.C.; Huang, G.; Mitchell, A.E. Comparison of amygdalin and benzaldehyde levels in California almond (Prunus dulcis) varietals. Acta Hortic. ISHS Proc. VII Int. Symp. Almonds Pistachios 2018, 1219, 1–7. [Google Scholar] [CrossRef]
- Dudareva, N.; Pichersky, E.; Gershenzon, J. Biochemistry of Plant Volatiles. Plant Physiol. 2004, 135, 1993–2011. [Google Scholar] [CrossRef] [Green Version]


| Cultivar/Selection | 2012 (n) | 2015 (n) | Genotype a | Ref. | Bitterness b(0–10) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trees | Samples | Trees | Samples | |||||||
| 1 | IRTA-7 (Lauranne × OP c) | 8 | 17 | unknown | d | na e | ||||
| 2 | IRTA-9 (Masbovera × Lauranne) | 2 | 4 | SkSk | d | na | ||||
| 3 | IRTA-4 (A-202 × FGFP092) | 3 | 5 | 1 | 2 | SkSk | d | na | ||
| 4 | IRTA-10 (4-665 × Lauranne) | 2 | 4 | SkSk | d | 0 | ||||
| 5 | IRTA-12 (4-665 × Lauranne) | 3 | 6 | SkSk | d | na | ||||
| 6 | IRTA-11 (Primorskyi × Cristomorto) × IRTA-7) | 2 | 4 | unknown | d | na | ||||
| 7 | IRTA-8 (Anxaneta × IRTA-4) | 2 | 4 | SkSk | d | 0.7 | ||||
| 8 | Belona (Blanquerna × Belle d’Aurons) | 2 | 4 | SkSk | d | na | ||||
| 9 | Cambra (Ferragnes × Tuono f) | 2 | 4 | unknown | na | |||||
| 10 | Constantí (FGFD2 × OP) | 7 | 14 | SkSk | d | na | ||||
| 11 | Desmayo Largueta (Spanish local) | 2 | 4 | Sksk | [20] | 1.8 | ||||
| 12 | Felisia (Titan × Tuono) | 2 | 4 | unknown | d | na | ||||
| 13 | Ferragnes (Cristomorto × Aï) | 2 | 4 | 1 | 2 | SkSk | [12,20] | 0.5 | ||
| 14 | Francolí (Cristomorto × Tuono) | 7 | 13 | 1 | 2 | SkSk | [20] | 0 | ||
| 15 | Glorieta (Primorskiy × Cristomorto) | 6 | 14 | 1 | 2 | SkSk | [20] | 0 | ||
| 16 | Guara (syn. Tuono) | 6 | 11 | 1 | 2 | Sksk | [28] | 2.8 | ||
| 17 | Lauranne (Ferragnes × Tuono) | 2 | 4 | SkSk | [20] | 0.3 | ||||
| 18 | Marcona (Spanish local) | 2 | 4 | 1 | 2 | Sksk | [20] | 0.3 | ||
| 19 | Marinada (Lauranne × Glorieta) | 7 | 13 | 1 | 2 | SkSk | d | 0.2 | ||
| 20 | Marta (Ferragnes × Tuono) | 3 | 5 | Sksk | [17] | na | ||||
| 21 | Masbovera (Primorskiy × Cristomorto) | 6 | 11 | 1 | 2 | SkSk | [20] | 0.3 | ||
| 22 | Nonpareil (Californian reference) | 2 | 4 | Sksk | [17] | 1.1 | ||||
| 23 | Soleta (Blanquerna × Belle d’Aurons) | 5 | 10 | unknown | 0.4 | |||||
| 24 | Tarraco (FLTU18 × Anxaneta) | 6 | 12 | 1 | 2 | SkSk | d | 0.6 | ||
| 25 | Vairo (4-665 × Lauranne) | 6 | 12 | 1 | 2 | SkSk | d | 0.3 | ||
| 26 | IRTA-2 (A-60 × A-192) | 1 | 2 | SkSk | d | 0.6 | ||||
| 27 | IRTA-1 (Wawona × Lauranne) | 1 | 2 | Sksk | d | 0.3 | ||||
| 28 | IRTA-3 (4-665 × Lauranne) | 1 | 1 | SkSk | d | 0.6 | ||||
| 29 | 4-665 (Primorskiy × Cristomorto) | 1 | 2 | SkSk | [20] | 0 | ||||
| 30 | Cristomorto (Italian local) | 1 | 2 | SkSk | [20] | 0.4 | ||||
| 31 | Falsa Barese (Italian local) | 1 | 2 | Sksk | [20] | 1.3 | ||||
| 32 | FGFP092 (Ferragnes × Filippo Ceo) | 1 | 2 | Sksk | [20] | 0 | ||||
| 33 | FGTR13 (Ferragnes × Troito) | 1 | 2 | Sksk | [20] | 2.1 | ||||
| 34 | FLTU18 (Ferralise × Tuono) | 1 | 2 | Sksk | [20] | 0.3 | ||||
| 35 | Gabaix (Spanish local) | 1 | 2 | Sksk | [20] | 0.3 | ||||
| 36 | Garbí (Cristomorto × OP) | 1 | 2 | SkSk | [20] | 0.4 | ||||
| 37 | Genco (Italian local) | 1 | 2 | Sksk | [12,20] | 3.5 | ||||
| 38 | Primorskiy (Princess × Nikitskiy) | 1 | 2 | SkSk | [12,20] | 0.3 | ||||
| 39 | Ramillete (Spanish local) | 1 | 2 | SkSk | [12,20] | 0.4 | ||||
| 40 | Stelliete (Ferragnes × Tuono) | 1 | 2 | Sksk | [20] | 1.8 | ||||
| 41 | Tuono f (Italian local) | 1 | 2 | Sksk | [12,20] | 0.7 | ||||
| 42 | Bitter almond (Spanish feral) | 1 | 2 | sksk | d | 10 | ||||
| RT a (min) | Compound | T b (°C) | T c (min) | pH d | Sample e (g) |
|---|---|---|---|---|---|
| 6.36 | hexanal | 60 | 40 | 7 | ns f |
| 7.30 | 2-Methy-1-propanol | 40 | ns | ns | 1 |
| 7.98 | 2-Pentanol | 40 | ns | ns | ns |
| 10.17 | 1-Penten-3-ol | ns | 40 | ns | ns |
| 12.01 | 3-Methyl-1-butanol | ns | 40 | ns | 1 |
| 16.68 | 2-Methyl-3-buten-1-ol | 60 | 40 | 7 | ns |
| 17.95 | 1-Hexanol | 60 | 40 | 7 | ns |
| 19.23 | nonanal | 60 | 40 | ns | ns |
| 21.92 | 1-Heptanol | 60 | ns | ns | 1.5 |
| 24.10 | benzaldehyde | 60 | 40 | ns | ns |
| 32.69 | benzyl alcohol | 60 | 40 | ns | ns |
| 33.26 | phenylethyl alcohol | 60 | 40 | ns | ns |
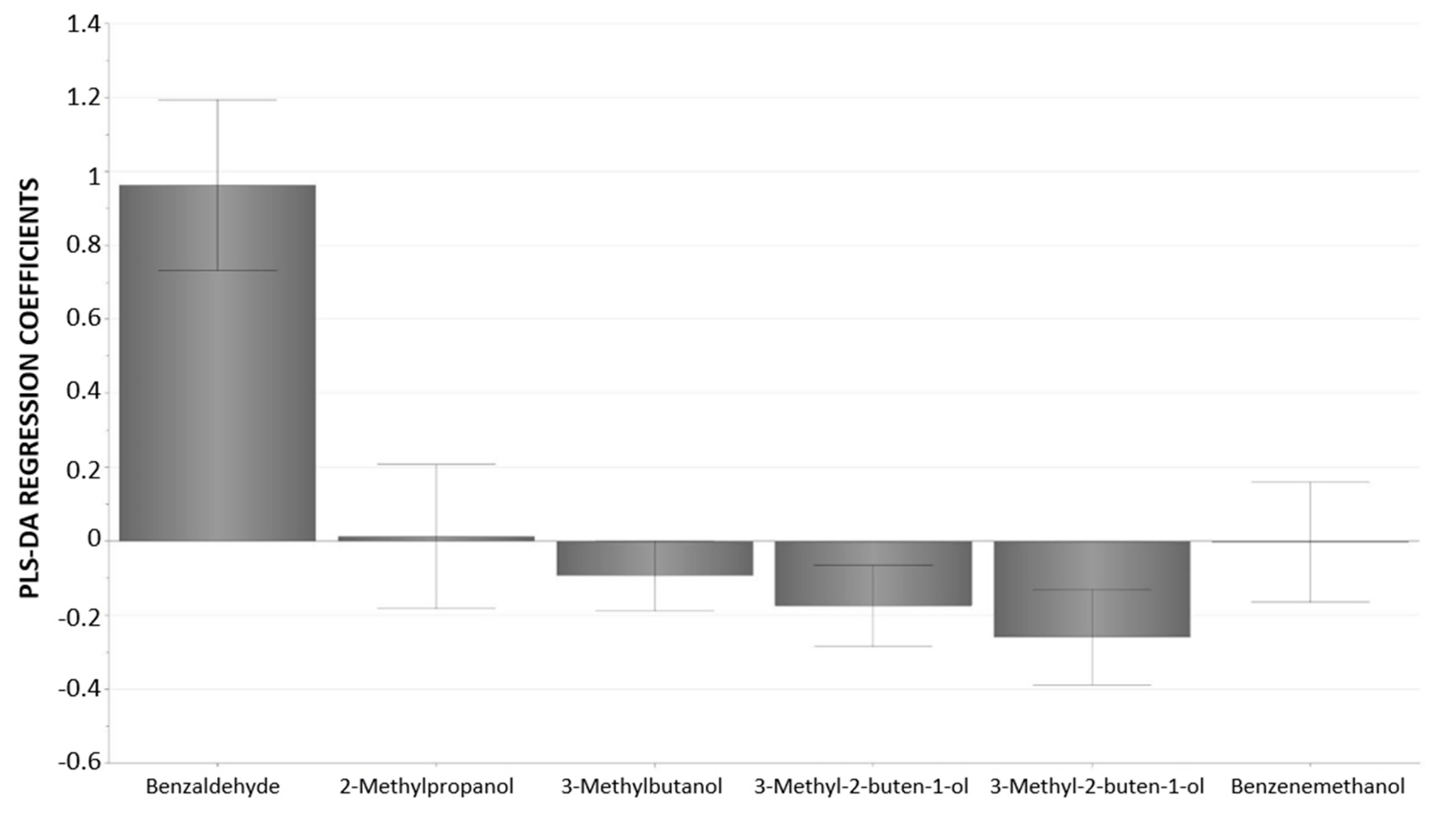
| Compound | Concentration a | t-Test b | Pearson Correlation c | |||
|---|---|---|---|---|---|---|
| SkSk (n = 153) | Sksk (n = 150) | sksk (n = 2) | p | r | p | |
| 2-Methylbutanal | 0.015 ± 0.011 | 0.007 ± 0.005 | 0.030 ± 0.000 | <0.001 | - | - |
| 3-Methylbutanal | 0.031 ± 0.019 | 0.013 ± 0.010 | 0.037 ± 0.004 | <0.001 | - | - |
| 2-Methylpropanol | 0.16 ± 0.15 | 0.070 ± 0.089 | 0.009 ± 0.000 | <0.001 | −0.236 | <0.001 |
| 1-Penten-3-ol | 0.092 ± 0.098 | 0.15 ± 0.15 | 0.011 ± 0.002 | <0.001 | - | - |
| 3-Methylbutan-1-ol | 0.91 ± 0.51 | 0.50 ± 0.44 | 0.034 ± 0.008 | <0.001 | −0.290 | <0.001 |
| 3-Methyl-3-buten-1-ol | 0.33 ± 0.15 | 0.23 ± 0.17 | 0.009 ± 0.001 | <0.001 | −0.213 | <0.01 |
| 3-Methyl-2-buten-1-ol | 0.29 ± 0.14 | 0.20 ± 0.13 | 0.012 ± 0.001 | <0.001 | −0.165 | <0.01 |
| benzaldehyde | 0.88 ± 1.06 | 26.3 ± 10.7 | 129.7 ± 4.7 | <0.001 | 1 | - |
| benzyl alcohol | 0.45 ± 0.31 | 1.29 ± 1.28 | 33.2 ± 2.5 | <0.001 | 0.767 | <0.001 |
| n | Correct Classification | SkSk | Sksk | |
|---|---|---|---|---|
| SkSk (homozygous) | 153 | 100% | 153 | 0 |
| Sksk (heterozygous) | 50 | 92% | 4 | 46 |
| Total | 203 | 98.03% | 157 | 46 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vichi, S.; Mayer, M.N.; León-Cárdenas, M.G.; Quintanilla-Casas, B.; Tres, A.; Guardiola, F.; Batlle, I.; Romero, A. Chemical Markers to Distinguish the Homo- and Heterozygous Bitter Genotype in Sweet Almond Kernels. Foods 2020, 9, 747. https://0-doi-org.brum.beds.ac.uk/10.3390/foods9060747
Vichi S, Mayer MN, León-Cárdenas MG, Quintanilla-Casas B, Tres A, Guardiola F, Batlle I, Romero A. Chemical Markers to Distinguish the Homo- and Heterozygous Bitter Genotype in Sweet Almond Kernels. Foods. 2020; 9(6):747. https://0-doi-org.brum.beds.ac.uk/10.3390/foods9060747
Chicago/Turabian StyleVichi, Stefania, Morgana N. Mayer, Maria G. León-Cárdenas, Beatriz Quintanilla-Casas, Alba Tres, Francesc Guardiola, Ignasi Batlle, and Agustí Romero. 2020. "Chemical Markers to Distinguish the Homo- and Heterozygous Bitter Genotype in Sweet Almond Kernels" Foods 9, no. 6: 747. https://0-doi-org.brum.beds.ac.uk/10.3390/foods9060747