Modeling of Filtration Processes—Microfiltration and Depth Filtration for Harvest of a Therapeutic Protein Expressed in Pichia pastoris at Constant Pressure
Abstract
:1. Introduction

1.1. Theory
1.2. Standard Blocking Model
1.3. Complete Blocking Model
1.4. Intermediate Blocking Model
1.5. Cake Filtration
1.6. Combined Models for Membrane Fouling
2. Experimental Section
2.1. Materials
2.2. Experimental Setup
2.3. Particle Size Distribution Measurement
| S.No. | Filter Type | Filter Name | Nominal Retention Rating (µm) | Description |
|---|---|---|---|---|
| 1. | Micro filters SuporGrade (Micro filter) | EKV | 0.2 | Filter Media is Supor EKV membrane (hydrophilic polyethersulfone) |
| 2. | Supra Cap 60 HP (Depth filter) | PDK5 | 1.5–20.0 | The HP-series depth filter sheets are comprised of two full thickness, graded, high-efficiency P-Series depth filter sheets in combination |
| 3. | Supra Cap P Series (Depth filter) | KS 50P | 0.4–0.8 | P series depth filter, combination of cellulose fibers, DE and perlite, pyrogen removal capability |
2.4. Turbidity Measurement
2.5. RP-HPLC
3. Results and Discussion



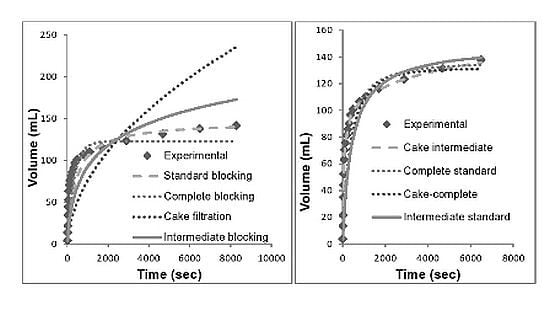
3.1. Direct Microfiltration (Option 1)
3.2. Centrifugation Followed by Microfiltration (Option 2)
| Model | Component mechanism | Equation | Parameters | Model |
|---|---|---|---|---|
| Cake-complete (6) | Cake filtration, complete blocking | Kc (s/m2), | Cake-complete (6) | |
| Cake-intermediate (7) | Cake filtration, intermediate blocking | Kc (s/m2), | Cake-intermediate (7) | |
| Complete-standard (8) | Complete blocking, standard blocking | Kb (s−1), | Complete-standard (8) | |
| Intermediate-standard (9) | Intermediate blocking, standard blocking | Ki (m−1), | Intermediate-standard (9) |
| Mechanism | EKV | PDK5 | PDH4 | PDE2 | EKMP | KS50P | EKSP | PDD1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameters | Error fit (SSR) | Parameters | Error fit (SSR) | Parameters | Error fit (SSR) | Parameters | Error fit (SSR) | Parameters | Error fit (SSR) | Parameters | Error fit (SSR) | Parameters | Error fit (SSR) | Parameters | Error fit (SSR) | |
| Standard blocking | KS = 87.98 | 169.17 | KS = 29.91 | 2611.7 | KS = 32.61 | 11,429 | KS = 41.91 | 3642.73 | KS = 45.84 | 5310.9 | KS = 32.61 | 11,429 | KS = 107.38 | 849.32 | KS = 50.63 | 1794.8 |
| Intermediate blocking | Ki = 188 | 174.97 | Ki = 84 | 2756.1 | Ki = 58 | 18,375 | Ki = 100 | 322.59 | Ki = 148 | 1460.5 | Ki = 58 | 18,735 | Ki = 297 | 36.07 | Ki = 100 | 534.43 |
| Complete blocking | Kb = 0.0358 | 1102.9 | Kb = 0.0194 | 8465.9 | Kb = 0.003 | 3295.9 | Kb = 0.0042 | 7255.56 | Kb = 0.0074 | 5947 | Kb = 0.003 | 9295.9 | Kb = 0.007 | 2075.2 | Kb = 0.0039 | 2115.39 |
| Cake filtration | Kc = 1.1 × 106 | 1337.3 | Kc = 7.02 × 105 | 32,325 | Kc = 1.0 × 106 | 43,477 | Kc = 3.49 × 106 | 1147.116 | Kc = 9.8 × 106 | 8372.8 | Kc = 1.45 × 106 | 43,477 | Kc = 2.6 × 107 | 587.15 | Kc = 3.0 × 106 | 1351.2 |
| Cake- Complete | Kb = 0.031 | 5.17 | Kb = 0.018 | 1393.3 | Kb = 0.0028 | 10,962 | Kb = 0.031 | 1059.88 | Kb = 0.007 | 1.45 × 103 | Kb = 0.0028 | 1.106 × 102 | Kb = 0.006 | 35.93 | Kb = 0.003 | 189.63 |
| Cake-intermediate | Kc = 2.08 × 105 | 294.96 | Ki = 50 | 1.23 × 103 | Ki = 45 | 1.8 × 102 | Ki = 35 | 3.66 | Kc = 9.7 × 104 | 393.59 | Kc = 1.15 × 104 | 31,831 | Kc = 2.6 × 106 | 37.87 | Kc = 1.28 × 104 | 297.81 |
| Complete-standard | Kb = 0.0034 | 147.41 | Kb = 0.017 | 2169.8 | Kb = 0.0026 | 10,098 | Kb = 0.0028 | 2673.53 | Kb = 0.007 | 4055.1 | Kb = 0.0026 | 10,091 | Kb = 0.006 | 672.45 | Kb = 0.0028 | 1930 |
| Intermediate-standard | Ki = 62 | 154.73 | Ki = 21 | 1241.8 | Ki = 20 | 12,040 | Ki = 50 | 2508.66 | Ki = 45 | 2544 | Ki = 20 | 12,101 | Ki = 150 | 419.28 | Ki = 50 | 896.01 |
3.3. Depth Filtration (Option 3)




3.4. Discussion
4. Conclusions
Nomenclature
| A | Available membrane frontal area (m2) |
| J0 | Initial flux (m/s) |
| Kb | Complete blocking constant (s−1) |
| Kc | Cake filtration constant (s/m2) |
| Ki | Intermediate blocking constant (m−1) |
| Ks | Standard blocking constant (m−1) |
| T | Time (s) |
| V | Volume filtered through available membrane area (m3/m2) |
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Cregg, J.M.; Vedvick, T.S.; Raschke, W.C. Recent advances in the expression of foreign genes in Pichiapastoris. Nat. Biotechnol. 1993, 11, 905–910. [Google Scholar] [CrossRef]
- Cregg, J.M.; Cereghino, J.L.; Shi, J.; Higgins, D.R. Recombinant protein expression in Pichiapastoris. Mol. Biotechnol. 2000, 16, 23–52. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Kuwae, S.; Ohya, T.; Ohda, T.; Ohyama, M.; Ohi, H.; Ohmura, T. High-level expression of recombinant human serum albumin from the methylotrophic yeast Pichia pastoris with minimal protease production and activation. J. Biosci. Bioeng. 2000, 89, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.R. Recombinant expression systems in the pharmaceutical industry. Appl. Microbiol. Biotechnol. 2004, 65, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Inan, M.; Meagher, M.M. Fermentation strategies for recombinant protein expression in the methylotrophic yeast Pichia pastoris. Biotechnol. Bioprocess Eng. 2000, 5, 275–287. [Google Scholar] [CrossRef]
- Van Reis, R.; Zydney, A. Bioprocess membrane technology. J. Membr. Sci. 2007, 297, 16–50. [Google Scholar] [CrossRef]
- Wang, A.; Lewus, R.; Rathore, A.S. Comparison of different options for harvest of a therapeutic protein product from high cell density yeast fermentation broth. Biotechnol. Bioeng. 2006, 94, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Russell, E.; Wang, A.; Rathore, A.S. Harvest of a Therapeutic Protein Product from High Cell Density Fermentation Broths. In Process Scale Bioseparations for the Biopharmaceutical Industry; CRC Press: Boca Raton, FL, USA, 2006; pp. 1–58. [Google Scholar]
- Charcosset, C. Membrane processes in biotechnology: An overview. Biotechnol. Adv. 2006, 24, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.S.; Shirke, A. Recent Developments in Membrane-Based Separations in Biotechnology Processes—Review. Prep. Biochem. Biotechnol. 2011, 41, 307–315. [Google Scholar] [CrossRef]
- Carrere, H.; Blaszkowa, F.; Roux de Balmann, H. Modelling the microfiltration of lactic acid fermentation broths and comparison of operating modes. Desalination 2002, 145, 201–206. [Google Scholar] [CrossRef] [Green Version]
- Grenier, A.; Meireles, M.; Aimar, P.; Carvin, P. Analysing flux decline in dead-end filtration. Chem. Eng. Res. Des. 2008, 86, 1281–1293. [Google Scholar] [CrossRef] [Green Version]
- Yuan, W.; Kocic, A.; Zydney, A.L. Analysis of humic acid fouling during microfiltration using a pore blockage-cake filtration model. J. Membr. Sci. 2002, 198, 51–62. [Google Scholar] [CrossRef]
- Rathore, A.S.; Kumar, V.; Arora, A.; Lute, S.; Brorson, K.; Shukla, A. Mechanistic modeling of viral filtration. J. Membr. Sci. 2014, 458, 96–103. [Google Scholar] [CrossRef]
- Hermia, J. Constant pressure blocking filtration laws—Application to power-law non-Newtonian fluids. Trans. I Chem E. 1982, 60, 183–187. [Google Scholar]
- Hale, M.B.; Daniels, W.F. A preliminary study of depth filtration at a constant rate. J. Biochem. Microb. Technol. Eng. 1961, 3, 139–150. [Google Scholar] [CrossRef]
- Guell, C.; Czekaj, P.; Davis, R.H. Microfiltration of protein mixtures and the effects of yeast on membrane fouling. J. Membr. Sci. 1999, 155, 113–122. [Google Scholar] [CrossRef]
- Ho, C.C.; Zydney, A.L. A combined pore blockage and cake filtration model for protein fouling during microfiltration. J. Colloid. Interf. Sci. 2000, 232, 389–399. [Google Scholar] [CrossRef]
- Velasco, C.; Ouammou, M.; Calvo, J.I.; Hernandez, A. Protein fouling in microfiltration: Deposition mechanism as a function of pressure for different pH. J. Colloid. Interf. Sci. 2003, 266, 148–152. [Google Scholar] [CrossRef]
- Schick, K. Enhancing dead end filtration throughput using a non-traditional liquid handling procedure. Filtr. Sep. 2003, 40, 30–33. [Google Scholar] [CrossRef]
- Kim, J.; DiGiano, F.A. Fouling models for low-pressure membrane systems. Sep. Purif. Technol. 2009, 68, 293–304. [Google Scholar] [CrossRef]
- Ochirkhuyag, B.; Mori, T.; Katsuoka, T.; Satone, H.; Tsubaki, J.; Choi, H.; Sugimoto, T. Development of a high-performance cake-less continuous filtration system. Chem. Eng. Sci. 2008, 63, 5274–5282. [Google Scholar] [CrossRef]
- Hong, S.; Krishna, P.; Hobbs, C.; Kim, D.; Cho, J. Variations in backwash efficiency during colloidal filtration of hollow-fiber microfiltration membranes. Desalination 2005, 173, 257–268. [Google Scholar] [CrossRef]
- Pegel, A.; Ubele, F.; Reiser, S.; Muller, D.; Dudziak, G. Evaluation of disposable systems for harvesting high cell density fed batch processes. BMC Proceedings 2011, 5. [Google Scholar] [CrossRef]
- Bowen, W.R.; Calvo, J.I.; Hernandez, A. Steps of membrane blocking in flux decline during protein microfiltration. J. Membr. Sci. 1995, 101, 153–165. [Google Scholar] [CrossRef]
- Bolton, G.; LaCasse, D.; Kuriyel, R. Combined models of membrane fouling: Development and application to microfiltration and ultrafiltration of biological fluids. J. Membr. Sci. 2006, 277, 75–84. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampath, M.; Shukla, A.; Rathore, A.S. Modeling of Filtration Processes—Microfiltration and Depth Filtration for Harvest of a Therapeutic Protein Expressed in Pichia pastoris at Constant Pressure. Bioengineering 2014, 1, 260-277. https://0-doi-org.brum.beds.ac.uk/10.3390/bioengineering1040260
Sampath M, Shukla A, Rathore AS. Modeling of Filtration Processes—Microfiltration and Depth Filtration for Harvest of a Therapeutic Protein Expressed in Pichia pastoris at Constant Pressure. Bioengineering. 2014; 1(4):260-277. https://0-doi-org.brum.beds.ac.uk/10.3390/bioengineering1040260
Chicago/Turabian StyleSampath, Muthukumar, Anupam Shukla, and Anurag S. Rathore. 2014. "Modeling of Filtration Processes—Microfiltration and Depth Filtration for Harvest of a Therapeutic Protein Expressed in Pichia pastoris at Constant Pressure" Bioengineering 1, no. 4: 260-277. https://0-doi-org.brum.beds.ac.uk/10.3390/bioengineering1040260