Multilocus Genotyping of Giardia duodenalis Occurring in Korean Native Calves
Abstract
:1. Introduction
2. Materials and Methods
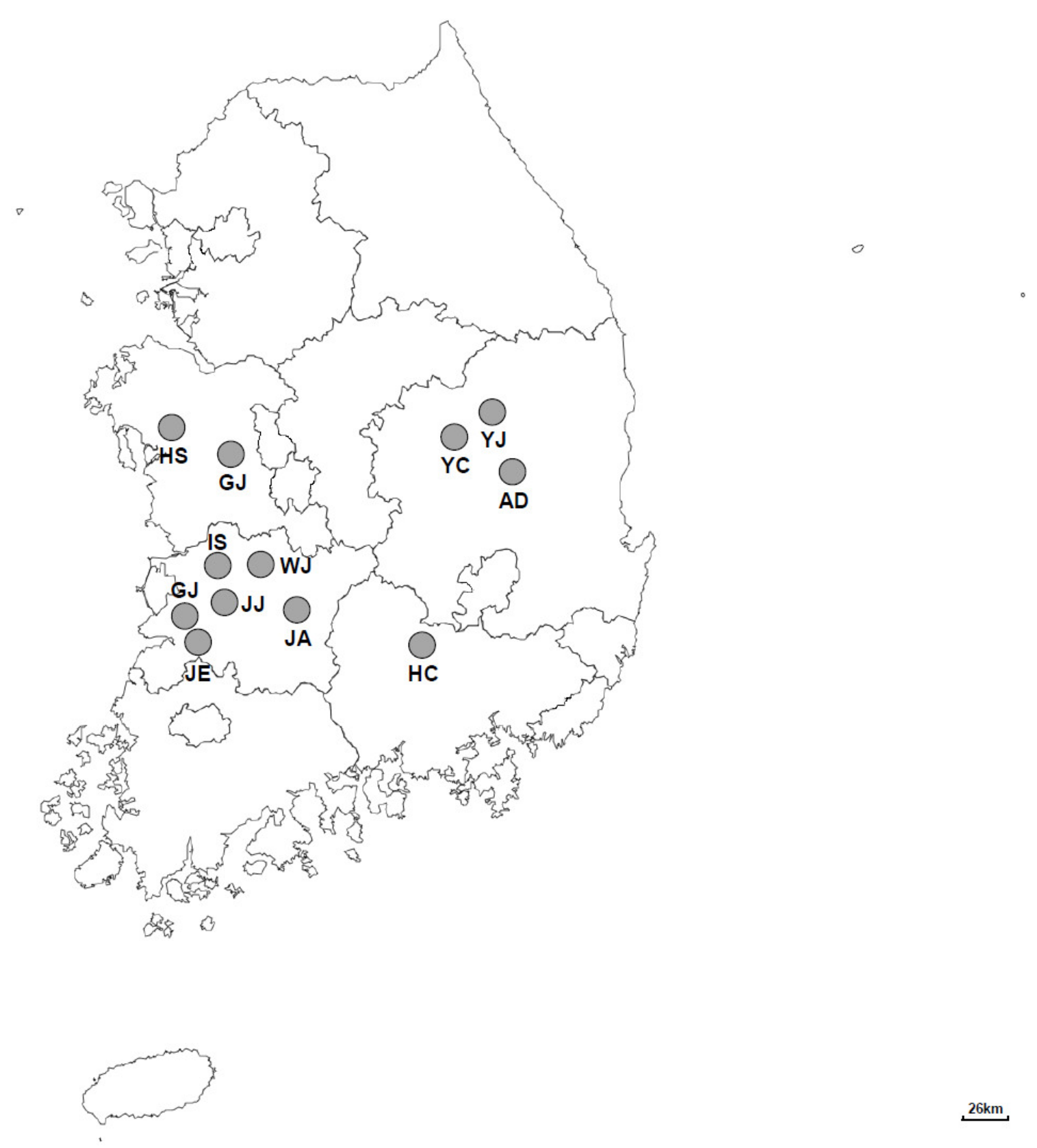
2.1. Sample Collection
2.2. DNA Extraction and Polymerase Chain Reaction (PCR) Amplification
2.3. Sequencing and Phylogenetic Analysis
2.4. Statistical Analysis
3. Results
3.1. Prevalence and Risk Factors of G. duodenalis
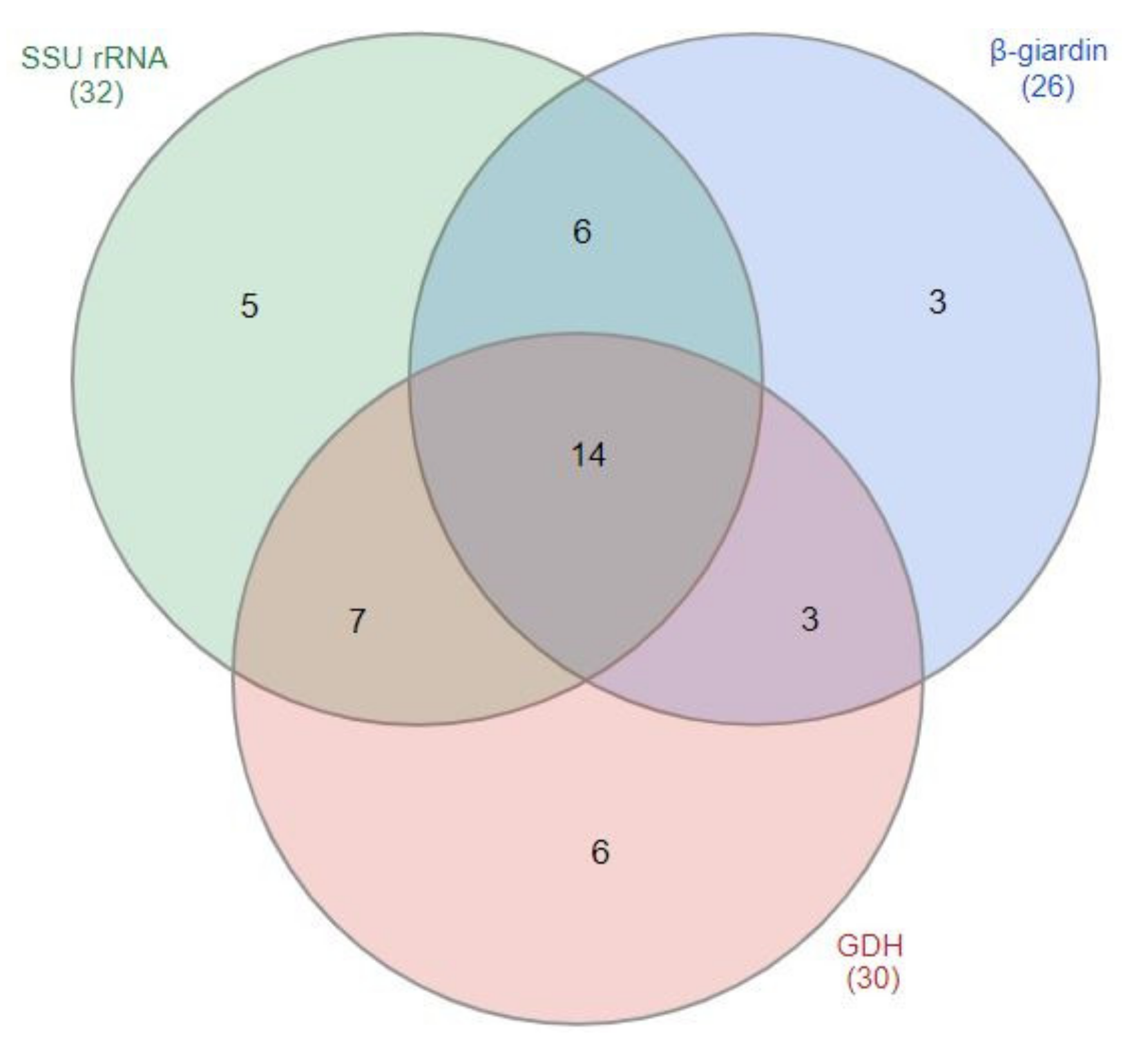
3.2. Detection of G. duodenalis by the Amplification of ssu, gdh, and bg
3.3. Nucleotide Sequencing
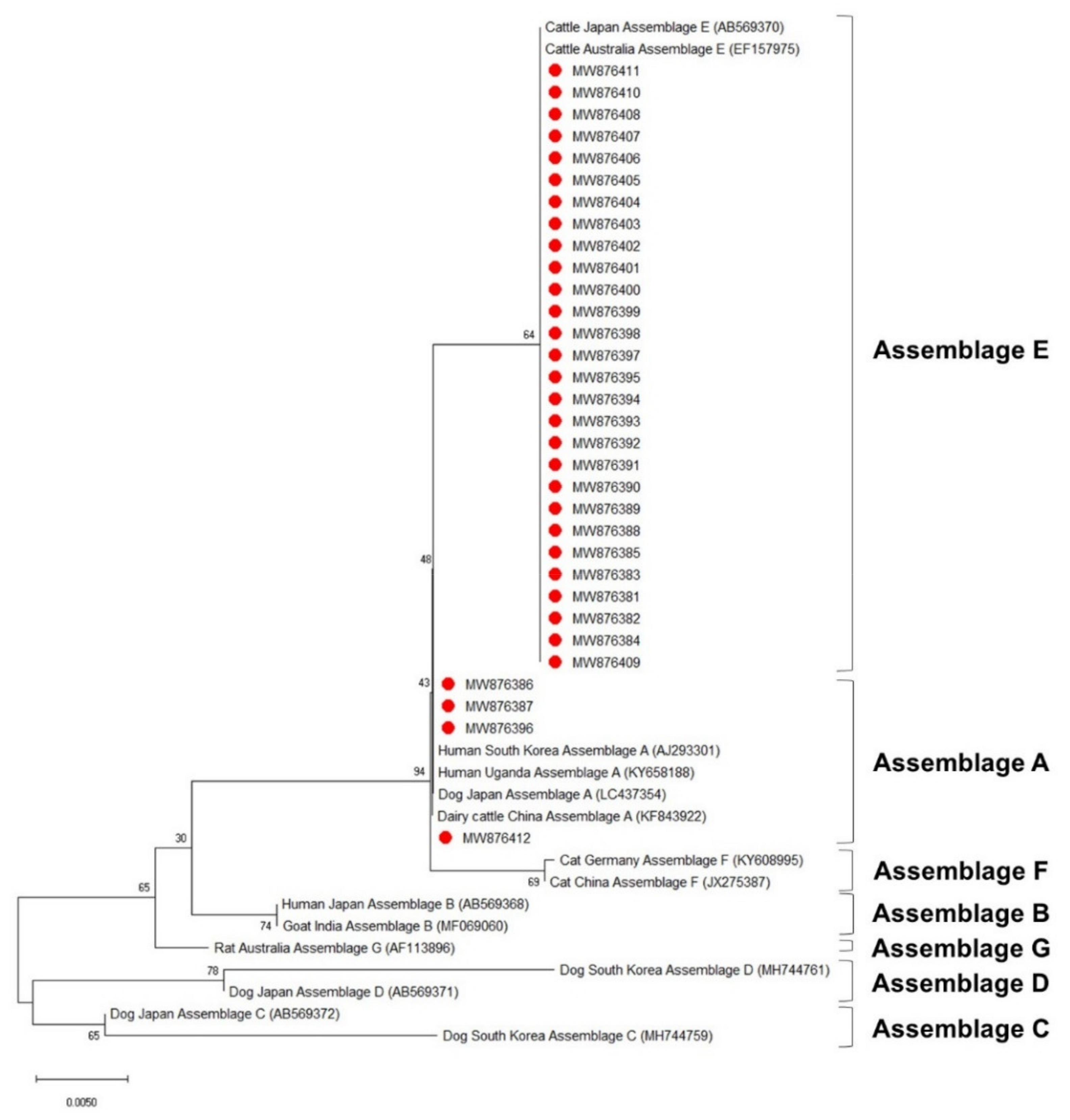
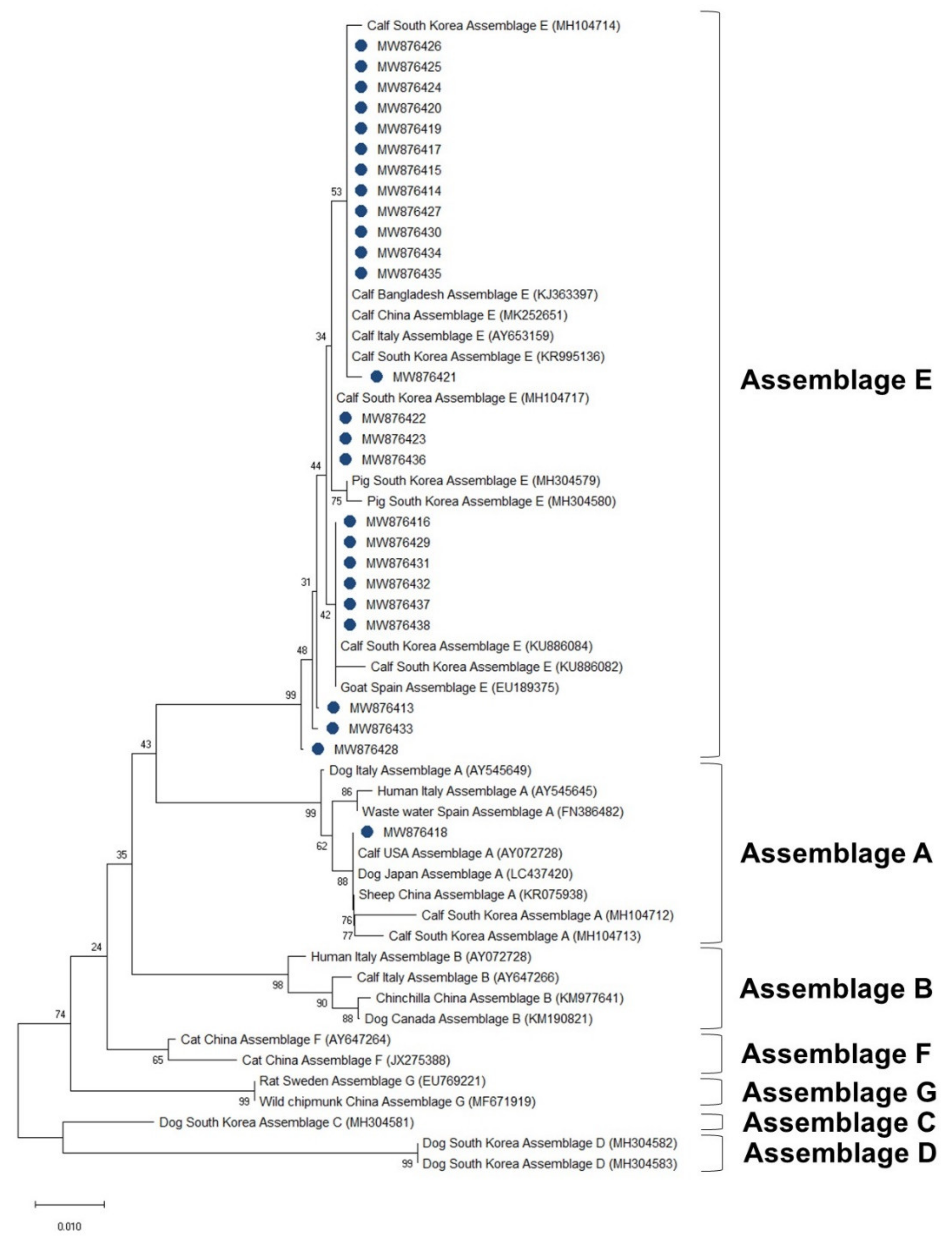
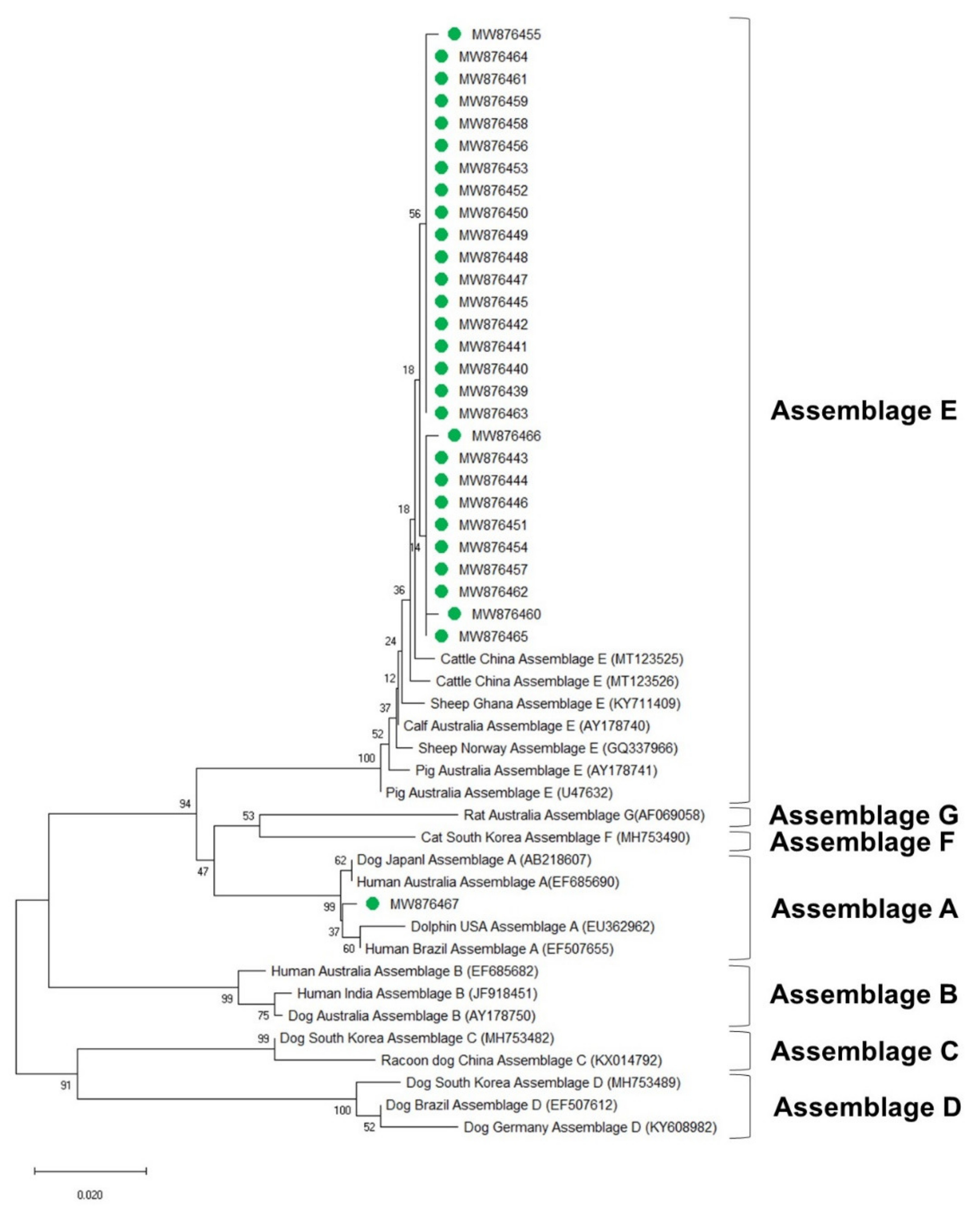
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheun, H.-I.; Kim, C.-H.; Cho, S.-H.; Ma, D.-W.; Goo, B.-L.; Na, M.-S.; Youn, S.-K.; Lee, W.-J. The first outbreak of giardiasis with drinking water in Korea. Osong Public Health Res. Perspect. 2013, 4, 89–92. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Xiao, L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011, 24, 110–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koloren, Z.; Seferoğlu, O.; Karanis, P. Occurency of Giardia duodenalis assemblages in river water sources of Black Sea, Turkey. Acta Trop. 2016, 164, 337–344. [Google Scholar] [CrossRef]
- Thompson, R.A. The zoonotic significance and molecular epidemiology of Giardia and giardiasis. Vet. Parasitol. 2004, 126, 15–35. [Google Scholar] [CrossRef]
- Buret, A.G. Mechanisms of epithelial dysfunction in giardiasis. Gut 2007, 56, 316–317. [Google Scholar] [CrossRef] [PubMed]
- Geurden, T.; Vercruysse, J.; Claerebout, E. Is Giardia a significant pathogen in production animals? Exp. Parasitol. 2010, 124, 98–106. [Google Scholar] [CrossRef]
- Bartelt, L.A.; Sartor, R.B. Advances in understanding Giardia: Determinants and mechanisms of chronic sequelae. F1000Prime Rep. 2015, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Heyworth, M.F. Giardia duodenalis genetic assemblages and hosts. Parasite 2016, 23, 13. [Google Scholar] [CrossRef] [Green Version]
- Smith, H.V.; Cacciò, S.M.; Tait, A.; McLauchlin, J.; Thompson, R.A. Tools for investigating the environmental transmission of Cryptosporidium and Giardia infections in humans. Trends Parasitol. 2006, 22, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Monis, P.T.; Caccio, S.M.; Thompson, R.A. Variation in Giardia: Towards a taxonomic revision of the genus. Trends Parasitol. 2009, 25, 93–100. [Google Scholar] [CrossRef]
- Khan, S.M.; Debnath, C.; Pramanik, A.K.; Xiao, L.; Nozaki, T.; Ganguly, S. Molecular evidence for zoonotic transmission of Giardia duodenalis among dairy farm workers in West Bengal, India. Vet. Parasitol. 2011, 178, 342–345. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; VanBik, D.; Kim, H.; Cho, A.; Kim, J.; Byun, J.; Oem, J.; Oh, S.; Kwak, D. Prevalence and molecular characterisation of Giardia duodenalis in calves with diarrhoea. Vet. Rec. 2016, 178, 633. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Han, D.-G.; Ryu, J.-H.; Chae, J.-B.; Chae, J.-S.; Yu, D.-H.; Park, J.; Park, B.-K.; Kim, H.-C.; Choi, K.-S. Identification of zoonotic Giardia duodenalis in Korean native calves with normal feces. J. Parasitol. Res. 2018, 117, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Gillhuber, J.; Pallant, L.; Ash, A.; Thompson, R.A.; Pfister, K.; Scheuerle, M.C. Molecular identification of zoonotic and livestock-specific Giardia-species in faecal samples of calves in Southern Germany. Parasit. Vectors 2013, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Geurden, T.; Geldhof, P.; Levecke, B.; Martens, C.; Berkvens, D.; Casaert, S.; Vercruysse, J.; Claerebout, E. Mixed Giardia duodenalis assemblage A and E infections in calves. Int. J. Parasitol. 2008, 38, 259–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trout, J.M.; Santín, M.; Greiner, E.; Fayer, R. Prevalence of Giardia duodenalis genotypes in pre-weaned dairy calves. Vet. Parasitol. 2004, 124, 179–186. [Google Scholar] [CrossRef]
- Geurden, T.; Somers, R.; Thanh, N.; Vien, L.; Nga, V.; Giang, H.; Dorny, P.; Giao, H.; Vercruysse, J. Parasitic infections in dairy cattle around Hanoi, northern Vietnam. Vet. Parasitol. 2008, 153, 384–388. [Google Scholar] [CrossRef]
- Li, S.; Zou, Y.; Zhang, X.-L.; Wang, P.; Chen, X.-Q.; Zhu, X.-Q. Prevalence and Multilocus Genotyping of Giardia lamblia in Cattle in Jiangxi Province, China: Novel Assemblage E Subtypes Identified. Korean J. Parasitol. 2020, 58, 681. [Google Scholar] [CrossRef]
- Naguib, D.; El-Gohary, A.H.; Mohamed, A.A.; Roellig, D.M.; Arafat, N.; Xiao, L. Age patterns of Cryptosporidium species and Giardia duodenalis in dairy calves in Egypt. Parasitol. Int. 2018, 67, 736–741. [Google Scholar] [CrossRef]
- Inpankaew, T.; Jiyipong, T.; Thadtapong, N.; Kengradomkij, C.; Pinyopanuwat, N.; Chimnoi, W.; Jittapalapong, S. Prevalence and genotype of Giardia duodenalis in dairy cattle from northern and northeastern part of Thailand. Acta Parasitol. 2015, 60, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, G.; Chen, G.; Jian, F.; Zhang, S.; Feng, C.; Wang, R.; Zhu, J.; Dong, H.; Hua, J. Multilocus genotyping of Giardia duodenalis in dairy cattle in Henan, China. PLoS ONE 2014, 9, e100453. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.X.; Tan, Q.D.; Zhao, G.H.; Ma, J.G.; Zheng, W.B.; Ni, X.T.; Zhao, Q.; Zhou, D.H.; Zhu, X.Q. Prevalence, risk factors and multilocus genotyping of Giardia intestinalis in dairy cattle, Northwest China. J. Eukaryot. Microbiol. 2016, 63, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Windeyer, M.; Leslie, K.; Godden, S.M.; Hodgins, D.; Lissemore, K.; LeBlanc, S. Factors associated with morbidity, mortality, and growth of dairy heifer calves up to 3 months of age. Prev. Vet. Med. 2014, 113, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Kwak, D.; Seo, M.-G. Genetic Analysis of zoonotic gastrointestinal Protozoa and Microsporidia in shelter cats in South Korea. Pathogens 2020, 9, 894. [Google Scholar] [CrossRef] [PubMed]
- Barigye, R.; Dyer, N.W.; Newell, T.K.; Khaitsa, M.L.; Trout, J.M.; Santin, M.; Fayer, R. Molecular and immunohistochemical detection of assemblage E, Giardia duodenalis in scouring North Dakota calves. Vet. Parasitol. 2008, 157, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Muñoz, M.T.; Cámara-Badenes, C.; del Carmen Martínez-Herrero, M.; Dea-Ayuela, M.A.; Pérez-Gracia, M.T.; Fernández-Barredo, S.; Santín, M.; Fayer, R. Multilocus genotyping of Giardia duodenalis in lambs from Spain reveals a high heterogeneity. Res. Vet. Sci. 2012, 93, 836–842. [Google Scholar] [CrossRef]
- Cacciò, S.M.; Ryan, U. Molecular epidemiology of giardiasis. Mol. Biochem. Parasitol. 2008, 160, 75–80. [Google Scholar] [CrossRef]
- Mendonça, C.; Almeida, A.; Castro, A.; de Lurdes Delgado, M.; Soares, S.; da Costa, J.M.C.; Canada, N. Molecular characterization of Cryptosporidium and Giardia isolates from cattle from Portugal. Vet. Parasitol. 2007, 147, 47–50. [Google Scholar] [CrossRef]
- Abeywardena, H.; Jex, A.R.; Firestone, S.M.; McPhee, S.; Driessen, N.; Koehler, A.V.; Haydon, S.R.; von Samson-Himmelstjerna, G.; Stevens, M.A.; Gasser, R.B. Assessing calves as carriers of Cryptosporidium and Giardia with zoonotic potential on dairy and beef farms within a water catchment area by mutation scanning. Electrophoresis 2013, 34, 2259–2267. [Google Scholar] [CrossRef]
| Classification | No. Tested | PCR | ||||
|---|---|---|---|---|---|---|
| No. of Positive Samples (%) | 95% CI * | OR * | p-Value | |||
| Constant | - | - | - | - | 0.06 | <0.001 |
| Age | ≤2 weeks ** | 462 | 7 (1.5) | 0.4–2.6 | 0.084 | 0.006 |
| 3–4 weeks | 138 | 9 (6.5) | 2.4–10.6 | 0.471 | 0.399 | |
| 5–6 weeks | 107 | 15 (14.0) | 7.4–20.6 | 1.081 | 0.929 | |
| 7–8 weeks | 26 | 4 (15.4) | 1.5–29.3 | 1.045 | 0.965 | |
| 9–12 weeks | 46 | 7 (15.2) | 4.8–25.6 | 1.072 | 0.940 | |
| ≥12 weeks | 13 | 2 (15.4) | 0–35.0 | - | - | |
| Sex | Male | 402 | 23 (5.7) | 3.5–8.0 | <0.001 | 0.998 |
| Female | 373 | 21 (5.6) | 3.3–8.0 | 0.618 | 0.161 | |
| Unknown | 17 | - | - | - | - | |
| Season | Spring ** | 256 | 17 (6.7) | 3.6–9.7 | 6.446 | 0.018 |
| Summer | 320 | 19 (6.0) | 3.4–8.5 | 3.393 | 0.113 | |
| Autumn | 81 | 6 (7.4) | 1.7–13.1 | 5.016 | 0.062 | |
| Winter | 135 | 2 (1.5) | 0–3.5 | - | - | |
| Fecal type | Normal | 349 | 8 (2.3) | 0.7–3.9 | - | - |
| Pasty | 137 | 4 (2.9) | 0.1–5.7 | 1.332 | 0.654 | |
| Watery ** | 234 | 23 (9.8) | 6.0–13.6 | 4.319 | 0.001 | |
| Hemorrhagic | 72 | 9 (12.5) | 4.9–20.1 | 2.123 | 0.169 | |
| Total | 792 | 44 (5.6) | 4.0–7.2 | - | - | |
| Accession No. 1 | Nucleotides at Variable Positions | |||
|---|---|---|---|---|
| 45 | 150 | 252 | 393 | |
| MW876414, 15, 17, 19, 20, 24–27, 30, 34, 35 | T | A | T | T |
| MW876416, 29, 31, 32, 37, 38 | C | A | C | T |
| MW876422, 23, 36 | T | A | C | T |
| MW876413 | T | A | C | C |
| MW876421 | T | G | T | T |
| MW876428 | T | G | C | C |
| MW876433 | C | A | C | C |
| Accession No. 1 | Nucleotides at Variable Positions | |||
|---|---|---|---|---|
| 258 | 306 | 342 | 387 | |
| MW876439–42, 45, 47–50, 52, 53, 56, 58, 59, 61, 63, 64 | C | T | A | C |
| MW876443, 44, 46, 51, 54, 57, 62, 65 | C | C | A | C |
| MW876455 | A | T | A | C |
| MW876460 | C | C | G | C |
| MW876466 | C | C | A | T |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, S.-I.; Jung, S.-H.; Lee, H.-K.; Choe, C.; Hur, T.-Y.; So, K.-M. Multilocus Genotyping of Giardia duodenalis Occurring in Korean Native Calves. Vet. Sci. 2021, 8, 118. https://0-doi-org.brum.beds.ac.uk/10.3390/vetsci8070118
Oh S-I, Jung S-H, Lee H-K, Choe C, Hur T-Y, So K-M. Multilocus Genotyping of Giardia duodenalis Occurring in Korean Native Calves. Veterinary Sciences. 2021; 8(7):118. https://0-doi-org.brum.beds.ac.uk/10.3390/vetsci8070118
Chicago/Turabian StyleOh, Sang-Ik, Suk-Han Jung, Han-Kyoung Lee, Changyong Choe, Tai-Young Hur, and Kyoung-Min So. 2021. "Multilocus Genotyping of Giardia duodenalis Occurring in Korean Native Calves" Veterinary Sciences 8, no. 7: 118. https://0-doi-org.brum.beds.ac.uk/10.3390/vetsci8070118







