Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid Detection of Avian Influenza Virus H9N2 HA Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethic Statement
2.2. Generation of RNA Standard
2.3. Viral RNA Extraction
2.4. H9 RT-RPA Primers and Exo Probe
2.5. Optimization of H9 RT-RPA Conditions
2.6. H9 RT-RPA Analytical Sensitivity
2.7. H9 RT-RPA Analytical Specificity
2.8. Clinical Accuracy of H9 RT-RPA
2.9. Statistical Methods
3. Results
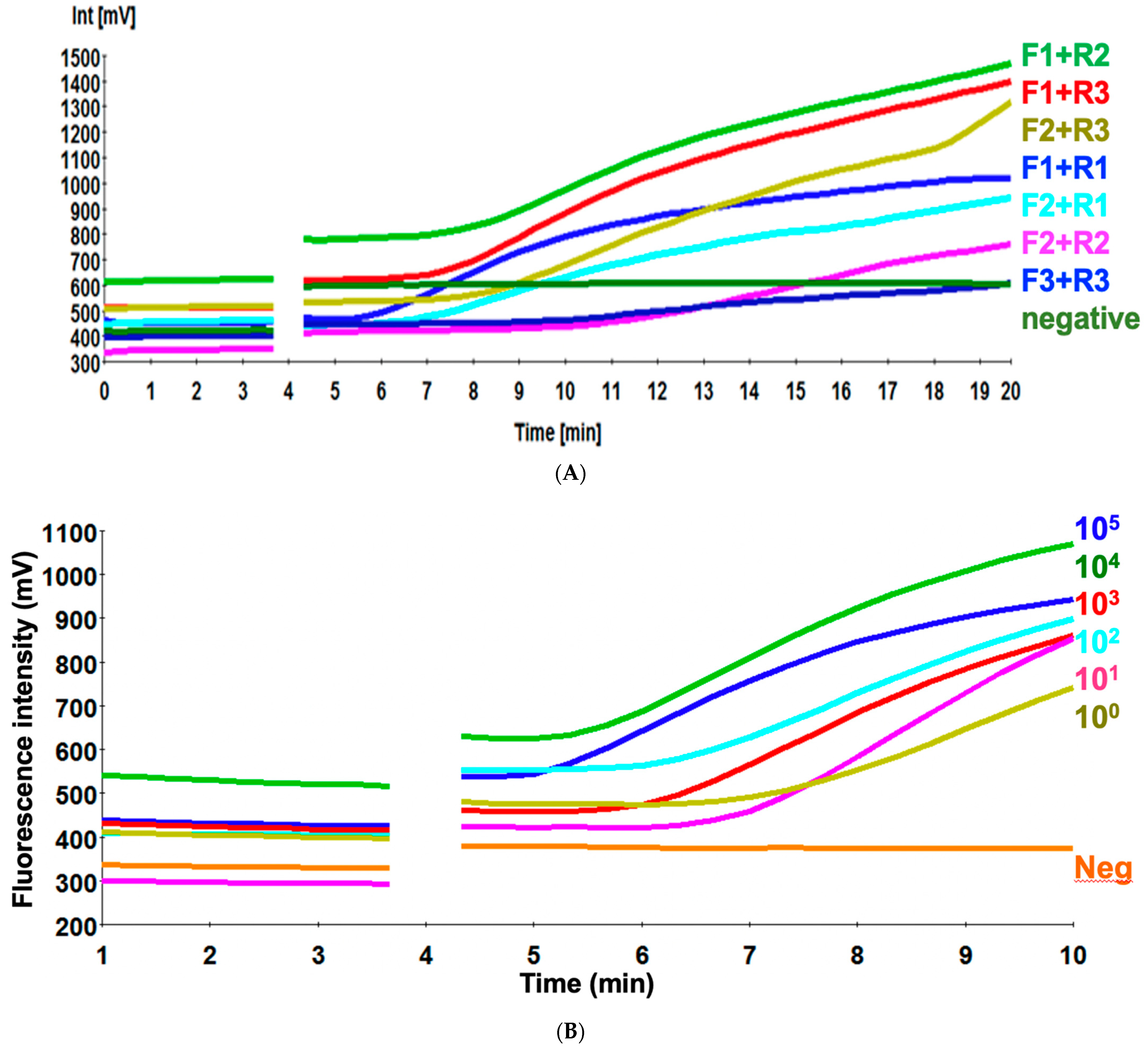
3.1. Screening of the Primer Sets
3.2. Sensitivity of H9 RT-RPA Assay
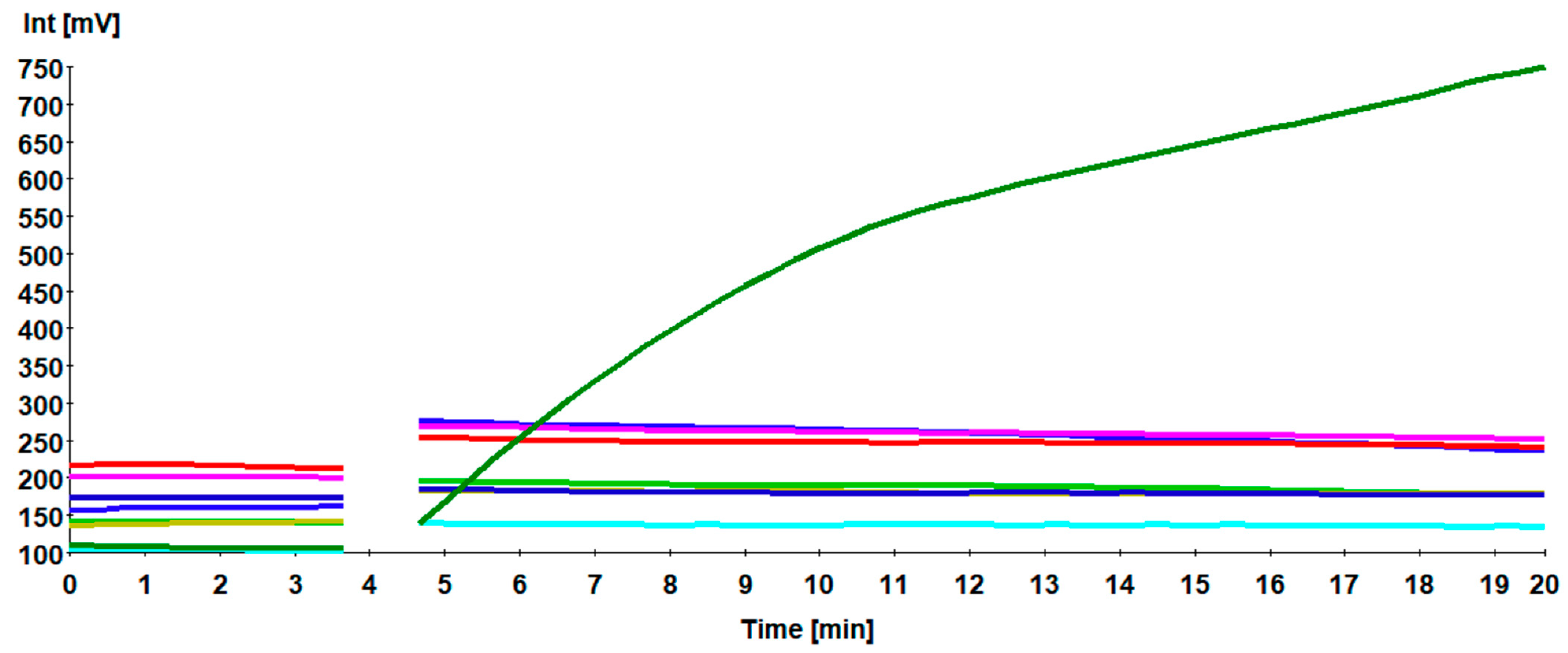
3.3. Specificity of H9 RT-RPA Assay
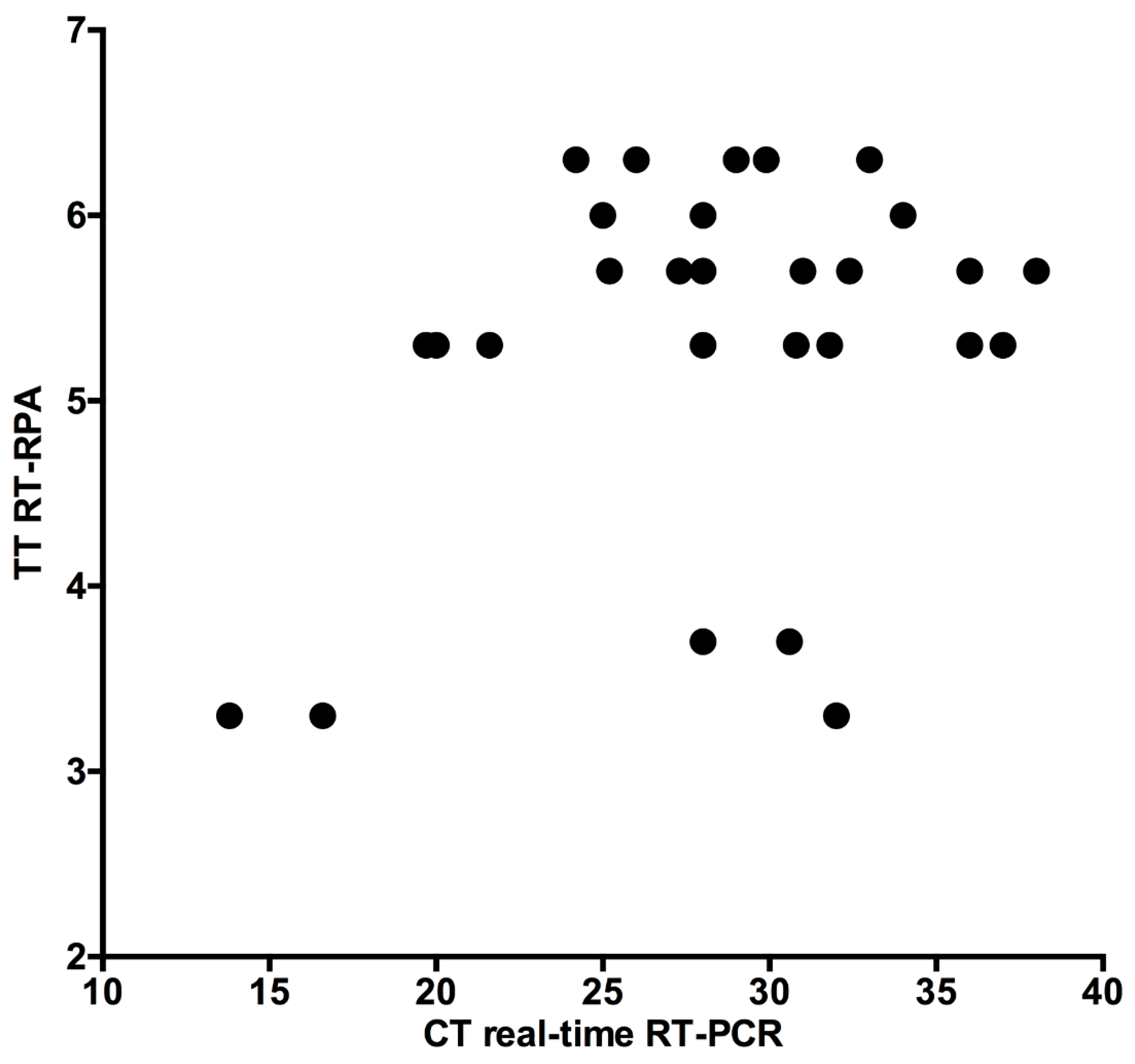
3.4. Clinical Accuracy of H9 RT-RPA Assay
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamb, R.A.; Krug, R.M. Orthomyxoviridae: The Viruses and Their Replication. In Fields Virology, 4th ed.; Knipe, D.M., Howley, P.M., Griffin, D.E., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001; pp. 1487–1531. [Google Scholar]
- Zheng, W.; Tao, Y.J. Structure and assembly of the influenza A virus ribonucleoprotein complex. FEBS Lett. 2013, 587, 1206–1214. [Google Scholar] [CrossRef] [Green Version]
- Amer, H.M.; Abd El Wahed, A.; Shalaby, M.A.; Almajhdi, F.N.; Hufert, F.T.; Weidmann, M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J. Virol. Methods 2013, 193, 337–340. [Google Scholar] [CrossRef]
- Peacock, T.H.P.; James, J.; Sealy, J.E.; Iqbal, M. A Global Perspective on H9N2 Avian Influenza Virus. Viruses 2019, 11, 620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taubenberger, J.K.; Kash, J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7, 440–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, S.; Zhang, Y.; Li, X.; Bo, H.; Wei, H.; Dong, L.; Yang, L.; Dong, J.; Liu, J.; Shu, Y.; et al. Molecular characterization and receptor binding specificity of H9N2 avian influenza viruses based on poultry-related environmental surveillance in China between 2013 and 2016. Virology 2019, 529, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Kandeil, A.; Hicks, J.T.; Young, S.G.; El Taweel, A.N.; Kayed, A.S.; Moatasim, Y.; Kutkat, O.; Bagato, O.; McKenzie, P.P.; Cai, Z.; et al. Active surveillance and genetic evolution of avian influenza viruses in Egypt, 2016–2018. Emerg. Microbes Infect. 2019, 8, 1370–1382. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H. Challenge for One Health: Co-Circulation of Zoonotic H5N1 and H9N2 Avian Influenza Viruses in Egypt. Viruses 2018, 10, 121. [Google Scholar] [CrossRef] [Green Version]
- Monne, I.; Hussein, H.A.; Fusaro, A.; Valastro, V.; Hamoud, M.M.; Khalefa, R.A.; Dardir, S.N.; Radwan, M.I.; Capua, I.; Cattoli, G. H9N2 influenza A virus circulates in H5N1 endemically infected poultry population in Egypt. Influenza Other Respir. Viruses 2013, 7, 240–243. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.P.; Peng, L.; Chen, L.; Jiang, L.F.; Zhang, Z.J.; Xiong, C.L.; Zhao, G.M.; Chen, Y.; Jiang, Q.W. Avian influenza viruses (AIVs) H9N2 are in the course of reassorting into novel AIVs. J. Zhejiang Univ. Sci. B 2018, 19, 409–414. [Google Scholar] [CrossRef] [PubMed]
- RahimiRad, S.; Alizadeh, A.; Alizadeh, E.; Hosseini, S.M. The avian influenza H9N2 at avian-human interface: A possible risk for the future pandemics. J. Res. Med. Sci. 2016, 21, 51. [Google Scholar] [CrossRef]
- Kandeil, A.; El-Shesheny, R.; Maatouq, A.; Moatasim, Y.; Cai, Z.; McKenzie, P.; Webby, R.; Kayali, G.; Ali, M.A. Novel reassortant H9N2 viruses in pigeons and evidence for antigenic diversity of H9N2 viruses isolated from quails in Egypt. J. Gen. Virol. 2017, 98, 548–562. [Google Scholar] [CrossRef]
- Li, H.; Liu, X.; Chen, F.; Zuo, K.; Wu, C.; Yan, Y.; Chen, W.; Lin, W.; Xie, Q. Avian Influenza Virus Subtype H9N2 Affects Intestinal Microbiota, Barrier Structure Injury, and Inflammatory Intestinal Disease in the Chicken Ileum. Viruses 2018, 10, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaleel, S.; Younus, M.; Idrees, A.; Arshad, M.; Khan, A.U.; Ehtisham-Ul-Haque, S.; Zaheer, M.I.; Tanweer, M.; Towakal, F.; Munibullah; et al. Pathological Alterations in Respiratory System During Co-infection with Low Pathogenic Avian Influenza Virus (H9N2) and Escherichia Coli in Broiler Chickens. J. Vet. Res. 2017, 61, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Naguib, M.M.; Arafa, A.S.; Parvin, R.; Beer, M.; Vahlenkamp, T.; Harder, T.C. Insights into genetic diversity and biological propensities of potentially zoonotic avian influenza H9N2 viruses circulating in Egypt. Virology 2017, 511, 165–174. [Google Scholar] [CrossRef]
- Abdelwhab, E.M.; Abdel-Moneim, A.S. Epidemiology, ecology and gene pool of influenza A virus in Egypt: Will Egypt be the epicentre of the next influenza pandemic? Virulence 2015, 6, 6–18. [Google Scholar] [CrossRef] [Green Version]
- OIE. Avian Influenza in Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; Office International Des Epizooties: Paris, France, 2008; pp. 465–481. [Google Scholar]
- OIE. Avian Influenza, Chapter 3.3.4 Terrestrial Manual; Office International Des Epizooties: Paris, France, 2018; pp. 821–841. [Google Scholar]
- Yehia, N.; Arafa, A.S.; Abd El Wahed, A.; El-Sanousi, A.A.; Weidmann, M.; Shalaby, M.A. Development of reverse transcription recombinase polymerase amplification assay for avian influenza H5N1 HA gene detection. J. Virol. Methods 2015, 223, 45–49. [Google Scholar] [CrossRef]
- Abd El Wahed, A.; El-Deeb, A.; El-Tholoth, M.; Abd El Kader, H.; Ahmed, A.; Hassan, S.; Hoffmann, B.; Haas, B.; Shalaby, M.A.; Hufert, F.T.; et al. A portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus. PLoS ONE 2013, 8, e71642. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Li, X.; Peng, B.; Wu, W.; Wang, X.; Liu, H.; Yuan, L.; Fang, S.; Lu, J. Rapid Detection of Avian Influenza A Virus (H7N9) by Lateral Flow Dipstick Recombinase Polymerase Amplification. Biol. Pharm. Bull. 2018, 41, 1804–1808. [Google Scholar] [CrossRef] [Green Version]
- Sun, N.; Wang, W.; Wang, J.; Yao, X.; Chen, F.; Li, X.; Yinglei, Y.; Chen, B. Reverse transcription recombinase polymerase amplification with lateral flow dipsticks for detection of influenza A virus and subtyping of H1 and H3. Mol. Cell. Probes 2018, 42, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.K.; Chen, C.C.; Lin, H.J.; Lin, H.Y.; Chen, T.T.; Wang, L.C. Combination of multiplex reverse transcription recombinase polymerase amplification assay and capillary electrophoresis provides high sensitive and high-throughput simultaneous detection of avian influenza virus subtypes. J. Vet. Sci. 2020, 21, e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Yang, P.P.; Zhang, Y.H.; Tian, K.Y.; Bian, C.Z.; Zhao, J. Development of a reverse transcription recombinase polymerase amplification combined with lateral-flow dipstick assay for avian influenza H9N2 HA gene detection. Transbound. Emerg. Dis. 2019, 66, 546–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Euler, M.; Wang, Y.; Heidenreich, D.; Patel, P.; Strohmeier, O.; Hakenberg, S.; Niedrig, M.; Hufert, F.T.; Weidmann, M. Development of a panel of recombinase polymerase amplification assays for detection of biothreat agents. J. Clin. Microbiol. 2013, 51, 1110–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Zeng, F.; Cong, F.; Huang, B.; Zhu, Y.; Wu, M.; Xu, F.; Yuan, W.; Huang, R.; Guo, P. Development and evaluation of a broadly reactive reverse transcription recombinase polymerase amplification assay for rapid detection of murine norovirus. BMC Vet. Res. 2018, 14, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishida, N.; Sakoda, Y.; Eto, M.; Sunaga, Y.; Kida, H. Co-infection of Staphylococcus aureus or Haemophilus paragallinarum exacerbates H9N2 influenza A virus infection in chickens. Arch. Virol. 2004, 149, 2095–2104. [Google Scholar] [CrossRef]
- Slomka, M.J.; Hanna, A.; Mahmood, S.; Govil, J.; Krill, D.; Manvell, R.J.; Shell, W.; Arnold, M.E.; Banks, J.; Brown, I.H. Phylogenetic and molecular characteristics of Eurasian H9 avian influenza viruses and their detection by two different H9-specific RealTime reverse transcriptase polymerase chain reaction tests. Vet. Microbiol. 2013, 162, 530–542. [Google Scholar] [CrossRef] [Green Version]
- Parikh, R.; Mathai, A.; Parikh, S.; Chandra Sekhar, G.; Thomas, R. Understanding and using sensitivity, specificity and predictive values. Indian J. Ophthalmol. 2008, 56, 45–50. [Google Scholar] [CrossRef]
- El Wahed, A.A.; Patel, P.; Maier, M.; Pietsch, C.; Rüster, D.; Böhlken-Fascher, S.; Kissenkötter, J.; Behrmann, O.; Frimpong, M.; Diagne, M.M.; et al. Suitcase Lab for Rapid Detection of SARS-CoV-2 Based on Recombinase Polymerase Amplification Assay. Anal. Chem. 2021, 93, 2627–2634. [Google Scholar] [CrossRef]
- Chen, H.T.; Zhang, J.; Sun, D.H.; Ma, L.N.; Liu, X.T.; Cai, X.P.; Liu, Y.S. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J. Virol. Methods 2008, 151, 200–203. [Google Scholar] [CrossRef]
- Lee, J.; Cho, A.Y.; Ko, H.H.; Ping, J.F.; Ma, L.J.; Chai, C.F.; Noh, J.Y.; Jeong, J.; Jeong, S.; Kim, Y.; et al. Evaluation of insulated isothermal PCR devices for the detection of avian influenza virus. J. Virol. Methods 2021, 292, 114126. [Google Scholar] [CrossRef]
- Hu, M.; Jin, Y.; Zhou, J.; Huang, Z.; Li, B.; Zhou, W.; Ren, H.; Yue, J.; Liang, L. Genetic Characteristic and Global Transmission of Influenza A H9N2 Virus. Front. Microbiol. 2017, 8, 2611. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Boudreau, D.K.; Bergeron, M.G. Influence of sequence mismatches on the specificity of recombinase polymerase amplification technology. Mol. Cell. Probes 2015, 29, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Abd El Wahed, A.; Weidmann, M.; Hufert, F.T. Diagnostics-in-a-Suitcase: Development of a portable and rapid assay for the detection of the emerging avian influenza A (H7N9) virus. J. Clin. Virol. 2015, 69, 16–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Name | Sequence |
|---|---|
| H9 RPA-F1 | ATGGCTGCAGATAGGGATTCCACTCAAAAGGCAG |
| H9 RPA-F2 | TTCCACTCAAAAGGCAGTTGACAAAATAAC |
| H9 RPA-F3 | TTGGTATGGTTTCCAACATTCAAATGATC |
| H9 RPA-R1 | TAGCAACTCTGCATTATATGCCCATACAT |
| H9 RPA-R2 | TTCACGTTTGCGTCATGCTCATCGAGTGTTTTC |
| H9 RPA-R3 | ATGCTCATCGAGTGTTTTCTGGTTCTCAAG |
| H9-exo probe | CATGAATTCAGTGAGGTTGAAACTAGAC(BHQ1-dT) (tetrahydrofuran residue) (FAM-dT) ATGATCAATAACA-PH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yehia, N.; Eldemery, F.; Arafa, A.-S.; Abd El Wahed, A.; El Sanousi, A.; Weidmann, M.; Shalaby, M. Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid Detection of Avian Influenza Virus H9N2 HA Gene. Vet. Sci. 2021, 8, 134. https://0-doi-org.brum.beds.ac.uk/10.3390/vetsci8070134
Yehia N, Eldemery F, Arafa A-S, Abd El Wahed A, El Sanousi A, Weidmann M, Shalaby M. Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid Detection of Avian Influenza Virus H9N2 HA Gene. Veterinary Sciences. 2021; 8(7):134. https://0-doi-org.brum.beds.ac.uk/10.3390/vetsci8070134
Chicago/Turabian StyleYehia, Nahed, Fatma Eldemery, Abdel-Satar Arafa, Ahmed Abd El Wahed, Ahmed El Sanousi, Manfred Weidmann, and Mohamed Shalaby. 2021. "Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid Detection of Avian Influenza Virus H9N2 HA Gene" Veterinary Sciences 8, no. 7: 134. https://0-doi-org.brum.beds.ac.uk/10.3390/vetsci8070134







