Aspergillus Metabolome Database for Mass Spectrometry Metabolomics
Abstract
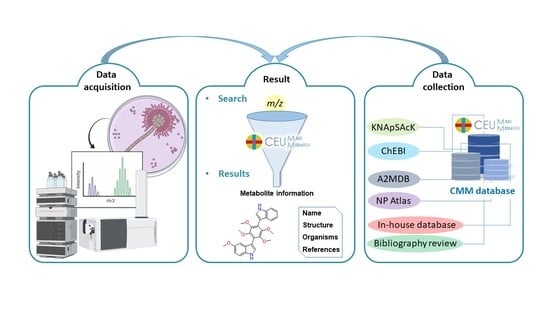
:1. Introduction
2. Materials and Methods
2.1. Collection and Integration of Aspergillus Metabolites
2.2. Creation of the Search Tool
3. Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Richardson, M.; Rautemaa-Richardson, R. Exposure to Aspergillus in home and healthcare facilities’ water environments: Focus on biofilms. Microorganisms 2019, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vadlapudi, V.; Borah, N.; Yellusani, K.R.; Gade, S.; Reddy, P.; Rajamanikyam, M.; Vempati, L.N.S.; Gubbala, S.P.; Chopra, P.; Upadhyayula, S.M.; et al. Aspergillus Secondary Metabolite Database, a resource to understand the Secondary metabolome of Aspergillus genus. Sci. Rep. 2017, 7, 7325–7334. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [Green Version]
- Perfect, J.R.; Cox, G.M.; Lee, J.Y.; Kauffman, C.A.; de Repentigny, L.; Chapman, S.W.; Morrison, V.A.; Pappas, P.; Hiemenz, J.W.; Stevens, D.A.; et al. The Impact of culture isolation of Aspergillus species: A hospital-based survey of Aspergillosis. Clin. Infect. Dis. 2001, 33, 1824–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, G.E.; Novak-Frazer, L. Chronic Pulmonary Aspergillosis—Where Are We? and Where Are We Going? J. Fungi 2016, 2, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latgé, J.P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. [Google Scholar] [CrossRef]
- Filho, A.P.D.C.; Brancini, G.T.P.; de Castro, P.A.; Valero, C.; Ferreira Filho, J.A.; Silva, L.P.; Rocha, M.C.; Malavazi, I.; de Moraes Pontes, J.G.; Fill, T.; et al. Aspergillus fumigatus G-protein coupled receptors gprm and gprj are important for the regulation of the cell wall integrity pathway, secondary metabolite production, and virulence. mBIO 2020, 11, e02458-20. [Google Scholar] [CrossRef]
- Guo, Y.; Kasahara, S.; Jhingran, A.; Tosini, N.L.; Zhai, B.; Aufiero, M.A.; Mills, K.A.M.; Gjonbalaj, M.; Espinosa, V.; Rivera, A.; et al. During Aspergillus infection, monocyte-derived DCs, neutrophils, and plasmacytoid dcs enhance innate immune defense through CXCR3-Dependent Crosstalk. Cell Host Microbe 2020, 28, 104–116. [Google Scholar] [CrossRef]
- Gourama, H.; Bullerman, L.B. Aspergillus flavus and Aspergillus parasiticus: Aflatoxigenic fungi of concern in foods and feeds: A review. J. Food Prot. 1995, 58, 1395–1404. [Google Scholar] [CrossRef]
- Perrone, G.; Susca, A.; Cozzi, G.; Ehrlich, K.; Varga, J.; Frisvad, J.C.; Meijer, M.; Noonim, P.; Mahakarnchanakul, W.; Samson, R.A. Biodiversity of Aspergillus species in some important agricultural products. Stud. Mycol. 2007, 59, 53–66. [Google Scholar] [CrossRef]
- Sheikh-Ali, S.I.; Ahmad, A.; Mohd-Setapar, S.; Zakaria, Z.A.; Abdul-Talib, N.; Khamis, A.K.; Hoque, M.E. The potential hazards of Aspergillus sp. in foods and feeds, and the role of biological treatment: A review. J. Microbiol. 2014, 52, 807–818. [Google Scholar] [CrossRef]
- Lin, S.; Schranz, J.; Teutsch, S.M. Aspergillosis case-fatality rate: Systematic review of the literature. Clin. Infect. Dis. 2001, 32, 358–366. [Google Scholar] [CrossRef]
- Sugui, J.A.; Kwon-Chung, K.J.; Juvvadi, P.R.; Steinbach, W.J.; Latgé, J.P. Aspergillus fumigatus and related species. Cold Spring Harb. Perspect. Med. 2014, 5, a019786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosmidis, C.; Denning, D.W. The clinical spectrum of pulmonary Aspergillosis. Thorax 2015, 70, 270–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tischler, B.Y.; Hohl, T.M. Menacing mold: Recent advances in Aspergillus pathogenesis and host defense. J. Mol. Biol. 2019, 431, 4229–4246. [Google Scholar] [CrossRef] [PubMed]
- Paulussen, C.; Hallsworth, J.E.; Álvarez-Pérez, S.; Nierman, W.C.; Hamill, P.G.; Blain, D.; Rediers, H.; Lievens, B. Ecology of aspergillosis: Insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 2017, 10, 296–322. [Google Scholar] [CrossRef] [Green Version]
- Zakaria, A.; Osman, M.; Dabboussi, F.; Rafei, R.; Mallat, H.; Papon, N.; Bouchara, J.; Hamze, M. Recent trends in the epidemiology, diagnosis, treatment, and mechanisms of resistance in clinical Aspergillus species: A general review with a special focus on the Middle Eastern and North African region. J. Infect. Public Health 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Brandt, P.; Garbe, E.; Vylkova, S. Catch the wave: Metabolomic Analyses in Human Pathogenic Fungi. PLoS Pathog. 2020, 16, e1008757. [Google Scholar] [CrossRef]
- Geiser, D.M. Sexual structures in Aspergillus: Morphology, importance and genomics. Med. Mycol. 2009, 47, S21–S26. [Google Scholar] [CrossRef]
- Ohkura, M.; Cotty, P.J.; Orbach, M.J. Comparative Genomics of Aspergillus flavus S and L Morphotypes Yield Insights into Niche Adaptation. G3 Genes Genom. Genet. 2018, 8, 3915–3930. [Google Scholar] [CrossRef] [Green Version]
- Sui, Y.; Ouyang, L.; Schütze, T.; Cheng, S.; Meyer, V.; Zhuang, Y. Comparative genomics of the aconidial Aspergillus niger strain LDM3 predicts genes associated with its high protein secretion capacity. Appl. Microbiol. Biotechnol. 2020, 104, 2623–2637. [Google Scholar] [CrossRef] [PubMed]
- Espindola, A.S.; Schneider, W.; Cardwell, K.F.; Carrillo, Y.; Hoyt, P.R.; Marek, S.M.; Melouk, H.A.; Garzon, C.D. Inferring the presence of aflatoxin-producing Aspergillus flavus strains using RNA sequencing and electronic probes as a transcriptomic screening tool. PLoS ONE 2018, 13, e0198575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Yuan, K.; Zhou, Q.; Lu, C.; Du, L.; Liu, F. Mechanism of antifungal activity of Perilla frutescens essential oil against Aspergillus flavus by transcriptomic analysis. Food Control 2020, 123, 107703. [Google Scholar] [CrossRef]
- Jiang, C.; Li, Z.; Shi, Y.; Guo, D.; Pang, B.; Chen, X.; Shao, D.; Liu, Y.; Shi, J. Bacillus subtilis inhibits Aspergillus carbonarius by producing iturin A, which disturbs the transport, energy metabolism, and osmotic pressure of fungal cells as revealed by transcriptomics analysis. Int. J. Food Microbiol. 2020, 330, 108783. [Google Scholar] [CrossRef] [PubMed]
- Shishodia, S.K.; Shankar, J. Proteomic analysis revealed ROS-mediated growth inhibition of Aspergillus terreus by shikonin. J. Proteom. 2020, 224, 103849. [Google Scholar] [CrossRef]
- Takagi, S.; Kojima, K.; Ohashi, S. Proteomic analysis on Aspergillus strains that are useful for industrial enzyme production. Biosci. Biotechnol. Biochem. 2020, 84, 2241–2252. [Google Scholar] [CrossRef]
- Deng, X.; Du, B.; Zhu, F.; Gao, Y.; Li, J. Proteomic analysis of Aspergillus niger 3.316 under heat stress. Microbiologyopen 2020, 9, e1012. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Lyu, G.; Luan, Y.; Yang, H.; Zhao, Z. Metabolomic study of the soybean pastes fermented by the single species Penicillium glabrum GQ1-3 and Aspergillus oryzae HGPA20. Food Chem. 2019, 295, 622–629. [Google Scholar] [CrossRef]
- Tawfike, A.F.; Romli, M.; Clements, C.; Abbott, G.; Young, L.; Schumacher, M.; Diederich, M.; Farag, M.; Edrada-Ebel, R. Isolation of anticancer and anti-trypanosome secondary metabolites from the endophytic fungus Aspergillus flocculus via bioactivity guided isolation and MS based metabolomics. J. Chromatogr. B 2019, 1106–1107, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Sun, X.; Feng, Q.; Luo, H.; Wassie, M.; Amee, M.; Amombo, E.; Chen, L. Comparative physiological and metabolomic analyses reveal mechanisms of Aspergillus aculeatus-mediated abiotic stress tolerance in tall fescue. Plant Physiol. Bioch. 2019, 142, 342–350. [Google Scholar] [CrossRef]
- Zhou, B.; Ma, C.; Ren, X.; Xia, T.; Li, X. LC-MS/MS-based metabolomic analysis of caffeine-degrading fungus Aspergillus sydowii during tea fermentation. J. Food Sci. 2020, 85, 477–485. [Google Scholar] [CrossRef]
- Romsdahl, J.; Blachowicz, A.; Chiang, A.J.; Chiang, Y.; Masonjones, S.; Yaegashi, J.; Countryman, S.; Karouia, F.; Kalkum, M.; Stajich, J.E.; et al. International Space Station conditions alter genomics, proteomics, and metabolomics in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2019, 103, 1363–1377. [Google Scholar] [CrossRef]
- Ma, Y.; Ling, T.-J.; Su, X.-Q.; Jiang, B.; Nian, B.; Chen, L.-J.; Liu, M.-L.; Zhang, Z.-Y.; Wang, D.-P.; Mu, Y.-Y.; et al. Integrated proteomics and metabolomics analysis of tea leaves fermented by Aspergillus niger, Aspergillus tamarii and Aspergillus fumigatus. Food Chem. 2021, 334, 127560. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics-the link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Oliver, S. The next wave in metabolome analysis. Trends Biotechnol. 2005, 23, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Raffler, J.; Römisch-Margl, W.; Petersen, A.-K.; Pagel, P.; Blöchl, F.; Hengstenberg, C.; Illig, T.; Meisinger, C.; Stark, K.; Wichmann, H.-E.; et al. Identification and MS-assisted interpretation of genetically influenced NMR signals in human plasma. Genome Med. 2013, 5, 13–27. [Google Scholar] [CrossRef] [Green Version]
- Blazenovic, I.; Kind, T.; Ji, J.; Fiehn, O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.H.; Nguyen, C.H.; Mamitsuka, H. Recent advances and prospects of computational methods for metabolite identification: A review with emphasis on machine learning approaches. Brief. Bioinform. 2018, 20, 2028–2043. [Google Scholar] [CrossRef]
- Monge, M.E.; Dodds, J.N.; Baker, E.S.; Edison, A.S.; Fernández, F.M. Challenges in identifying the dark molecules of life. Annu. Rev. Anal. Chem. 2019, 12, 177–199. [Google Scholar] [CrossRef]
- Gil-de-la-Fuente, A.; Armitage, E.G.; Otero, A.; Barbas, C.; Godzien, J. Differentiating signals to make biological sense—A guide through databases for MS-based non-targeted metabolomics. Electrophoresis 2017, 38, 2242–2256. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Afendi, F.M.; Parvin, A.K.; Ono, N.; Tanaka, K.; Morita, A.H.; Sato, T.; Sugiura, T.; Altaf-Ul-Amin, M.; Kanaya, S. KNApSAcK Metabolite activity database for retrieving the relationships between metabolites and biological activities. Plant Cell Physiol. 2014, 55, e7. [Google Scholar] [CrossRef] [Green Version]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016, 44, D1214–D1219. [Google Scholar] [CrossRef] [PubMed]
- Basenko, E.Y.; Pulman, J.A.; Shanmugasundram, A.; Harb, O.S.; Crouch, K.; Starns, D.; Warrenfeltz, S.; Aurrecoechea, C.; Stoeckert, J.C.J.; Kissinger, J.C.; et al. FungiDB: An integrated bioinformatic resource for fungi and oomycetes. J. Fungi 2018, 4, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerqueira, G.C.; Arnaud, M.B.; Inglis, D.O.; Skrzypek, M.S.; Binkley, G.; Simison, M.; Miyasato, S.R.; Binkley, J.; Orvis, J.; Shah, P.; et al. The Aspergillus genome database: Multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014, 42, D705–D710. [Google Scholar] [CrossRef] [Green Version]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The natural products atlas: An open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Djoumbou-Feunang, Y.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN—A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Bryant, S.H. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009, 37, W623–W633. [Google Scholar] [CrossRef]
- Gil-De-La-Fuente, A.; Godzien, J.; Saugar, S.; Garcia-Carmona, R.; Badran, H.; Wishart, D.S.; Barbas, C.; Otero, A. CEU Mass Mediator 3.0: A Metabolite Annotation Tool. J. Proteome Res. 2019, 18, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.-B.; Hubka, V.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Susca, A.; Tanney, J.B.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choque, E.; El-Rayess, Y.; Raynal, J.; Mathieu, F. Fungal naphtho-γ-pyrones—Secondary metabolites of industrial interest. Appl. Microbiol. Biotechnol. 2015, 99, 1081–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dotis, J.; Kondou, A.; Koukloumperi, E.; Karava, V.; Papadopoulou, A.; Gkogka, C.; Printza, N. Aspergillus peritonitis in peritoneal dialysis patients: A systematic review. J. Mycol. Med. 2020, 30, 101037. [Google Scholar] [CrossRef]
- Djoumbou-Feunang, Y.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform. 2016, 8, 61. [Google Scholar] [CrossRef] [Green Version]




| AspergillusDB | KEGG | LipidMaps | HMDB | METLIN |
|---|---|---|---|---|
| 2811 | 164 | 57 | 160 | 214 |
| AspergillusDB | NP Atlas | KNApSAcK | A2MDB |
|---|---|---|---|
| 2811 | 2029 | 621 | 807 |
| Main Class | Classyfire Node ID | Number of Compounds |
|---|---|---|
| Organooxygen compounds | CHEMONTID:0000323 | 430 |
| Organic oxides | CHEMONTID:0003940 | 378 |
| Oxacyclic compounds | CHEMONTID:0004140 | 245 |
| Carboxylic acids and derivatives | CHEMONTID:0000265 | 239 |
| Phenols | CHEMONTID:0000134 | 172 |
| Organonitrogen compounds | CHEMONTID:0000278 | 152 |
| Vinylogous acids | CHEMONTID:0003889 | 142 |
| Benzene and substituted derivatives | CHEMONTID:0002279 | 136 |
| Azacyclic compounds | CHEMONTID:0004139 | 136 |
| Heteroaromatic compounds | CHEMONTID:0004144 | 131 |
| Lactones | CHEMONTID:0000050 | 114 |
| Pyrans | CHEMONTID:0000086 | 91 |
| Benzopyrans | CHEMONTID:0000123 | 84 |
| Lactams | CHEMONTID:0000160 | 78 |
| Phenol ethers | CHEMONTID:0002341 | 73 |
| Dihydrofurans | CHEMONTID:0001983 | 54 |
| Fatty Acyls | CHEMONTID:0003909 | 49 |
| Indoles and derivatives | CHEMONTID:0000211 | 48 |
| Prenol lipids | CHEMONTID:0000259 | 43 |
| Naphthalenes | CHEMONTID:0000023 | 42 |
| Pyrroles | CHEMONTID:0000090 | 37 |
| Vinylogous esters | CHEMONTID:0003891 | 35 |
| Propargyl-type 1,3-dipolar organic compounds | CHEMONTID:0003633 | 33 |
| Naphthopyrans | CHEMONTID:0001640 | 32 |
| Pyrrolidines | CHEMONTID:0000218 | 29 |
| Carboximidic acids and derivatives | CHEMONTID:0002285 | 29 |
| Tetrahydrofurans | CHEMONTID:0002648 | 27 |
| Coumarans | CHEMONTID:0004189 | 25 |
| Epoxides | CHEMONTID:0000159 | 25 |
| Oxanes | CHEMONTID:0002012 | 25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil-de-la-Fuente, A.; Mamani-Huanca, M.; Stroe, M.C.; Saugar, S.; Garcia-Alvarez, A.; Brakhage, A.A.; Barbas, C.; Otero, A. Aspergillus Metabolome Database for Mass Spectrometry Metabolomics. J. Fungi 2021, 7, 387. https://0-doi-org.brum.beds.ac.uk/10.3390/jof7050387
Gil-de-la-Fuente A, Mamani-Huanca M, Stroe MC, Saugar S, Garcia-Alvarez A, Brakhage AA, Barbas C, Otero A. Aspergillus Metabolome Database for Mass Spectrometry Metabolomics. Journal of Fungi. 2021; 7(5):387. https://0-doi-org.brum.beds.ac.uk/10.3390/jof7050387
Chicago/Turabian StyleGil-de-la-Fuente, Alberto, Maricruz Mamani-Huanca, María C. Stroe, Sergio Saugar, Alejandra Garcia-Alvarez, Axel A. Brakhage, Coral Barbas, and Abraham Otero. 2021. "Aspergillus Metabolome Database for Mass Spectrometry Metabolomics" Journal of Fungi 7, no. 5: 387. https://0-doi-org.brum.beds.ac.uk/10.3390/jof7050387