LncRNAs in Cardiomyocyte Maturation: New Window for Cardiac Regenerative Medicine
Abstract
:1. Introduction
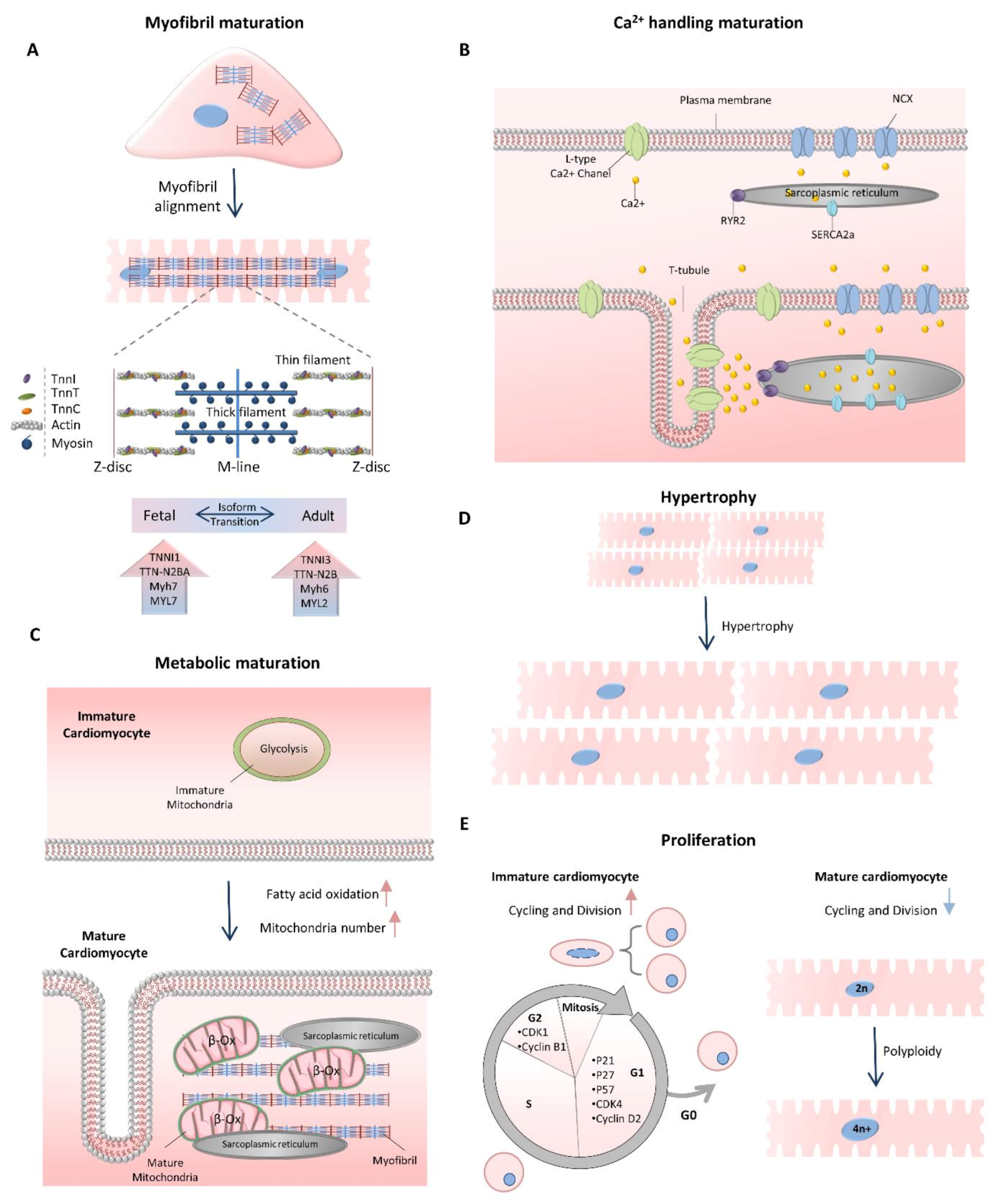
2. Cardiomyocyte Maturation
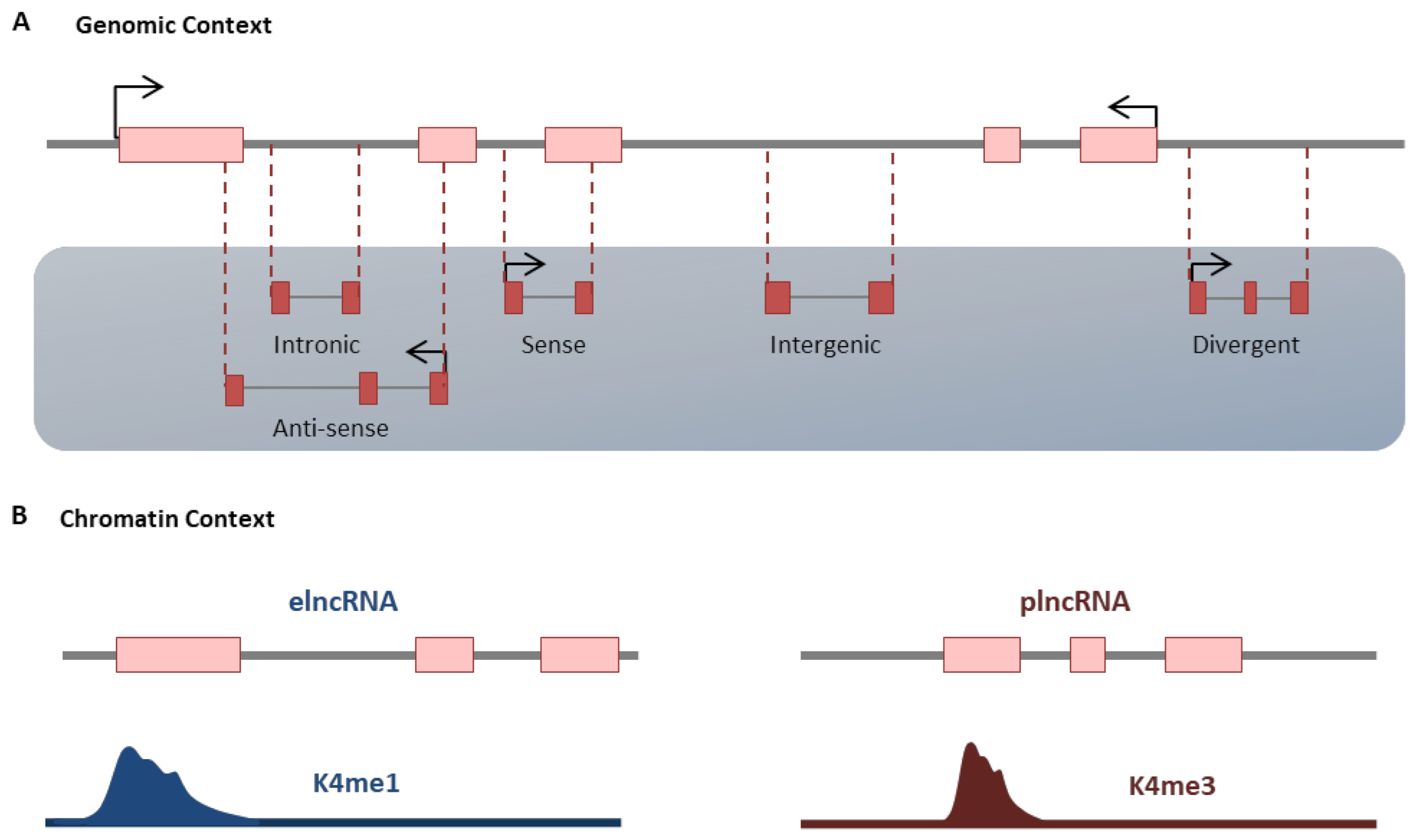
3. Noncoding RNAs
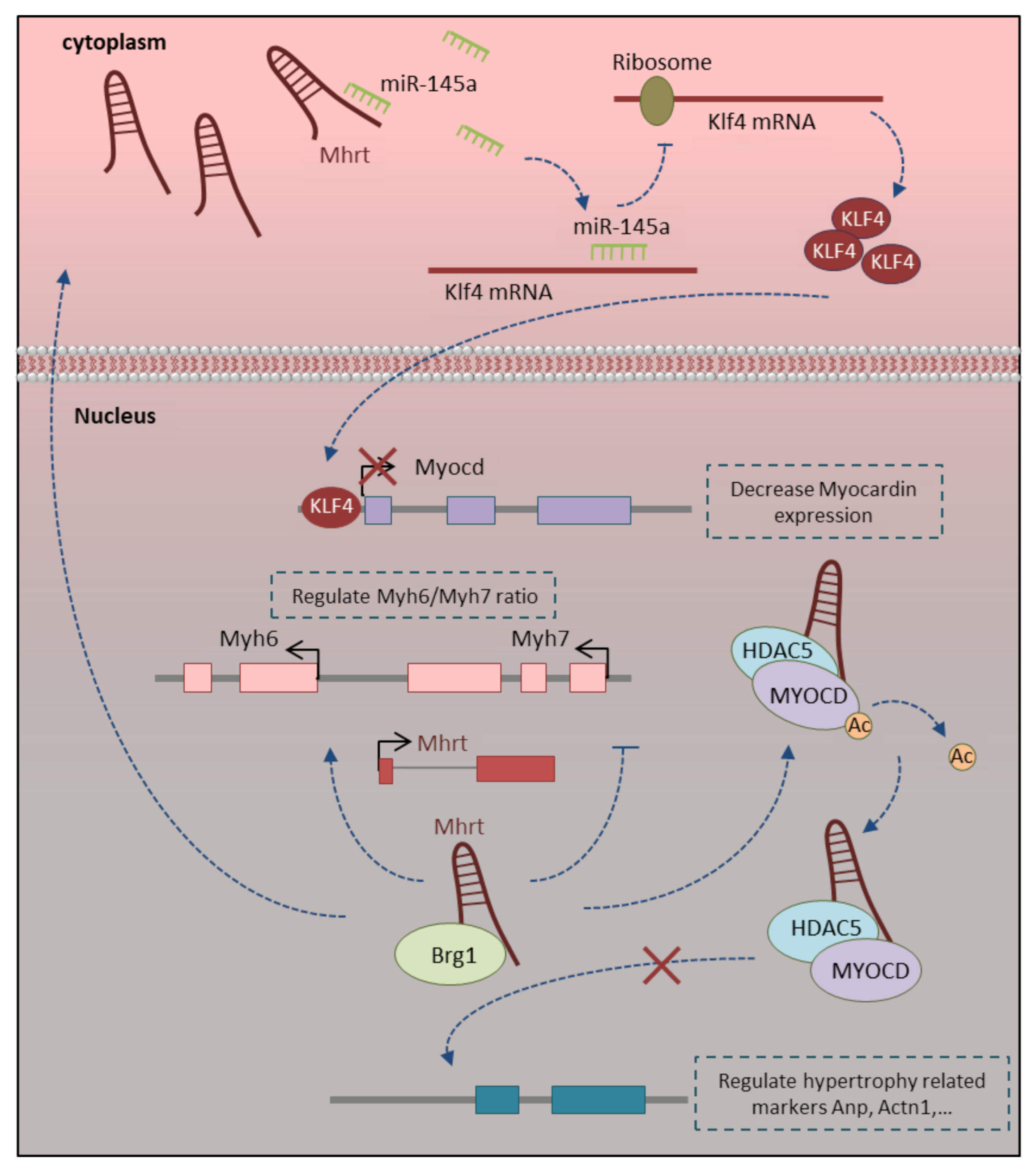
3.1. Mhrt-LncRNA
3.2. H19-LncRNA
3.3. Ahit LncRNA
3.4. Zfas1 and Dach1 LncRNAs
3.5. Carl and Mdrl LncRNAs
3.6. Cpr and Sarrah LncRNAs
3.7. Other Novel LncRNAs
4. Therapeutic Target of LncRNAs in Cardiac Remodeling
5. Conclusions
Funding
Conflicts of Interest
References
- Liu, Y.-W.; Chen, B.; Yang, X.; Fugate, J.A.; Kalucki, F.A.; Futakuchi-Tsuchida, A.; Couture, L.; Vogel, K.W.; Astley, C.A.; Baldessari, A.; et al. Human embryonic stem cell–derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat. Biotechnol. 2018, 36, 597–605. [Google Scholar] [CrossRef]
- Huang, C.Y.; Maia-Joca, R.P.M.; Ong, C.S.; Wilson, I.; DiSilvestre, D.; Tomaselli, G.F.; Reich, D.H. Enhancement of human iPSC-derived cardiomyocyte maturation by chemical conditioning in a 3D environment. J. Mol. Cell. Cardiol. 2020, 138, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef]
- Bektik, E.; Cowan, D.B.; Wang, D.-Z. Long non-coding RNAs in atrial fibrillation: Pluripotent stem cell-derived cardiomyocytes as a model system. Int. J. Mol. Sci. 2020, 21, 5424. [Google Scholar] [CrossRef]
- Karbassi, E.; Fenix, A.; Marchiano, S.; Muraoka, N.; Nakamura, K.; Yang, X.; Murry, C.E. Cardiomyocyte maturation: Advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 2020, 17, 341–359. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Pu, W.T. Cardiomyocyte maturation: New phase in development. Circ. Res. 2020, 126, 1086–1106. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Touma, M.; Kang, X.; Zhao, Y.; Cass, A.A.; Gao, F.; Biniwale, R.; Coppola, G.; Xiao, X.; Reemtsen, B.; Wang, Y. Decoding the long noncoding RNA during cardiac maturation: A roadmap for functional discovery. Circ. Cardiovasc. Genet. 2016, 9, 395–407. [Google Scholar] [CrossRef]
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzini, T.; Chang, C.-P.; Dorn, G.W., II; Thum, T.; Heymans, S. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015, 12, 415. [Google Scholar]
- Sizarov, A.; Ya, J.; de Boer, B.A.; Lamers, W.H.; Christoffels, V.M.; Moorman, A.F. Formation of the building plan of the human heart: Morphogenesis, growth, and differentiation. Circulation 2011, 123, 1125–1135. [Google Scholar] [CrossRef] [Green Version]
- Bird, S.; Doevendans, P.; Van Rooijen, M.; De La Riviere, A.B.; Hassink, R.; Passier, R.; Mummery, C. The human adult cardiomyocyte phenotype. Cardiovasc. Res. 2003, 58, 423–434. [Google Scholar] [CrossRef]
- Lundy, S.D.; Zhu, W.-Z.; Regnier, M.; Laflamme, M.A. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013, 22, 1991–2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iorga, B.; Schwanke, K.; Weber, N.; Wendland, M.; Greten, S.; Piep, B.; Dos Remedios, C.G.; Martin, U.; Zweigerdt, R.; Kraft, T.; et al. Differences in contractile function of myofibrils within human embryonic stem cell-derived cardiomyocytes vs. adult ventricular myofibrils are related to distinct sarcomeric protein isoforms. Front. Physiol. 2018, 8, 1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarkova, I.; Perriard, J.-C. The M-band: An elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005, 15, 477–485. [Google Scholar] [CrossRef]
- Reiser, P.J.; Portman, M.A.; Ning, X.-H.; Moravec, C.S. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am. J. Physiol. Heart Circ. Physiol. 2001, 280, H1814–H1820. [Google Scholar] [CrossRef] [Green Version]
- Siedner, S.; Krüger, M.; Schroeter, M.; Metzler, D.; Roell, W.; Fleischmann, B.K.; Hescheler, J.; Pfitzer, G.; Stehle, R. Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart. J. Physiol. 2003, 548, 493–505. [Google Scholar] [CrossRef]
- Mahdavi, V.; Lompre, A.; Chambers, A.; Nadal-Ginard, B. Cardiac myosin heavy chain isozymic transitions during development and under pathological conditions are regulated at the level of mRNA availability. Eur. Heart J. 1984, 5, 181–191. [Google Scholar] [CrossRef]
- Lahmers, S.; Wu, Y.; Call, D.R.; Labeit, S.; Granzier, H. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ. Res. 2004, 94, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Kubalak, S.W.; Miller-Hance, W.C.; O’Brien, T.X.; Dyson, E.; Chien, K.R. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J. Biol. Chem. 1994, 269, 16961–16970. [Google Scholar] [CrossRef]
- Louch, W.E.; Koivumäki, J.T.; Tavi, P. Calcium signalling in developing cardiomyocytes: Implications for model systems and disease. J. Physiol. 2015, 593, 1047–1063. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Gao, S.; Nie, L.; Tang, M.; Huang, W.; Luo, H.; Hu, X.; Xi, J.; Zhu, M.; Zheng, Y.; et al. Molecular and functional changes in voltage-gated Na+ channels in cardiomyocytes during mouse embryogenesis. Circ. J. 2011, 75, 2071–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veerman, C.C.; Mengarelli, I.; Lodder, E.M.; Kosmidis, G.; Bellin, M.; Zhang, M.; Dittmann, S.; Guan, K.; Wilde, A.A.; Schulze-Bahr, E.; et al. Switch From Fetal to Adult SCN 5A Isoform in Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes Unmasks the Cellular Phenotype of a Conduction Disease–Causing Mutation. J. Am. Heart Assoc. 2017, 6, e005135. [Google Scholar] [CrossRef]
- Goversen, B.; van der Heyden, M.A.; van Veen, T.A.; de Boer, T.P. The immature electrophysiological phenotype of iPSC-CMs still hampers in vitro drug screening: Special focus on IK1. Pharmacol. Ther. 2018, 183, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Boutjdir, M. Gene expression of SERCA2a and L-and T-type Ca channels during human heart development. Pediatr. Res. 2001, 50, 569–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bers, D.M. Cardiac excitation–contraction coupling. Nature 2002, 415, 198–205. [Google Scholar] [CrossRef]
- Peinkofer, G.; Burkert, K.; Urban, K.; Krausgrill, B.; Hescheler, J.; Saric, T.; Halbach, M. From early embryonic to adult stage: Comparative study of action potentials of native and pluripotent stem cell-derived cardiomyocytes. Stem Cells Dev. 2016, 25, 1397–1406. [Google Scholar] [CrossRef]
- Lieu, D.K.; Liu, J.; Siu, C.-W.; McNerney, G.P.; Tse, H.-F.; Abu-Khalil, A.; Huser, T.; Li, R.A. Absence of transverse tubules contributes to non-uniform ca2+ wavefronts in mouse and human embryonic stem cell–derived cardiomyocytes. Stem Cells Dev. 2009, 18, 1493–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, T.; Shaw, R.M. Cardiac T-tubule microanatomy and function. Physiol. Rev. 2017, 97, 227–252. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Pu, W.T. The architecture and function of cardiac dyads. Biophys. Rev. 2020, 12, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Jaswal, J.S. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 2010, 56, 130–140. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Dorn, G.W. Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ. Res. 2011, 109, 1327–1331. [Google Scholar] [CrossRef]
- Sim, C.B.; Ziemann, M.; Kaspi, A.; Harikrishnan, K.; Ooi, J.; Khurana, I.; Chang, L.; Hudson, J.E.; El-Osta, A.; Porrello, E.R. Dynamic changes in the cardiac methylome during postnatal development. FASEB J. 2015, 29, 1329–1343. [Google Scholar] [CrossRef] [Green Version]
- Uosaki, H.; Cahan, P.; Lee, D.I.; Wang, S.; Miyamoto, M.; Fernandez, L.; Kass, D.A.; Kwon, C. Transcriptional landscape of cardiomyocyte maturation. Cell Rep. 2015, 13, 1705–1716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaper, J.; Meiser, E.; Stämmler, G. Ultrastructural morphometric analysis of myocardium from dogs, rats, hamsters, mice, and from human hearts. Circ. Res. 1985, 56, 377–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seppet, E.K.; Kaambre, T.; Sikk, P.; Tiivel, T.; Vija, H.; Tonkonogi, M.; Sahlin, K.; Kay, L.; Appaix, F.; Braun, U.; et al. Functional complexes of mitochondria with Ca, MgATPases of myofibrils and sarcoplasmic reticulum in muscle cells. Biochim. Biophys. Acta BBA Bioenergy 2001, 1504, 379–395. [Google Scholar] [CrossRef] [Green Version]
- Dai, D.-F.; Danoviz, M.E.; Wiczer, B.; Laflamme, M.A.; Tian, R. Mitochondrial maturation in human pluripotent stem cell derived cardiomyocytes. Stem Cells Int. 2017, 2017, 5153625. [Google Scholar] [CrossRef]
- Yutzey, K.E. Neuregulin 1 makes heart muscle. Nature 2015, 520, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Grego-Bessa, J.; Luna-Zurita, L.; del Monte, G.; Bolós, V.; Melgar, P.; Arandilla, A.; Garratt, A.N.; Zang, H.; Mukouyama, Y.-S.; Chen, H.; et al. Notch signaling is essential for ventricular chamber development. Dev. Cell 2007, 12, 415–429. [Google Scholar] [CrossRef] [Green Version]
- Campa, V.M.; Gutiérrez-Lanza, R.; Cerignoli, F.; Díaz-Trelles, R.; Nelson, B.; Tsuji, T.; Barcova, M.; Jiang, W.; Mercola, M. Notch activates cell cycle reentry and progression in quiescent cardiomyocytes. J. Cell Biol. 2008, 183, 129–141. [Google Scholar] [CrossRef] [Green Version]
- Heallen, T.; Morikawa, Y.; Leach, J.; Tao, G.; Willerson, J.T.; Johnson, R.L.; Martin, J.F. Hippo signaling impedes adult heart regeneration. Development 2013, 140, 4683–4690. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Guo, H.; Cao, Y.; Zohrabian, S.; Zhou, P.; Ma, Q.; VanDusen, N.; Guo, Y.; Zhang, J.; Stevens, S.M.; et al. Acetylation of VGLL4 regulates Hippo-YAP signaling and postnatal cardiac growth. Dev. Cell 2016, 39, 466–479. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, T.M.; Ang, Y.-S.; Radzinsky, E.; Zhou, P.; Huang, Y.; Elfenbein, A.; Foley, A.; Magnitsky, S.; Srivastava, D. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell 2018, 173, 104–116.e112. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Meng, Q.; Yu, Y.; Shao, L.; Shen, Z. Adult Cardiomyocyte Proliferation: A New Insight for Myocardial Infarction Therapy. J. Cardiovasc. Transl. Res. 2020, 1–10. [Google Scholar] [CrossRef]
- Yatscoff, M.A.; Jaswal, J.S.; Grant, M.R.; Greenwood, R.; Lukat, T.; Beker, D.L.; Rebeyka, I.M.; Lopaschuk, G.D. Myocardial hypertrophy and the maturation of fatty acid oxidation in the newborn human heart. Pediatr. Res. 2008, 64, 643–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, M.; Barske, L.; Van Handel, B.; Rau, C.D.; Gan, P.; Sharma, A.; Parikh, S.; Denholtz, M.; Huang, Y.; Yamaguchi, Y.; et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet. 2017, 49, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- González-Rosa, J.M.; Sharpe, M.; Field, D.; Soonpaa, M.H.; Field, L.J.; Burns, C.E.; Burns, C.G. Myocardial polyploidization creates a barrier to heart regeneration in zebrafish. Dev. Cell 2018, 44, 433–446.e437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Zhang, C.-H.; Ammanamanchi, N.; Suresh, S.; Lewarchik, C.; Rao, K.; Uys, G.M.; Han, L.; Abrial, M.; Yimlamai, D.; et al. Control of cytokinesis by β-adrenergic receptors indicates an approach for regulating cardiomyocyte endowment. Sci. Transl. Med. 2019, 11, eaaw6419. [Google Scholar] [CrossRef]
- Peter, A.K.; Cheng, H.; Ross, R.S.; Knowlton, K.U.; Chen, J. The costamere bridges sarcomeres to the sarcolemma in striated muscle. Prog. Pediatric Cardiol. 2011, 31, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Paige, S.L.; Plonowska, K.; Xu, A.; Wu, S.M. Molecular regulation of cardiomyocyte differentiation. Circ. Res. 2015, 116, 341–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermij, S.H.; Abriel, H.; van Veen, T.A. Refining the molecular organization of the cardiac intercalated disc. Cardiovasc. Res. 2017, 113, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Epifantseva, I.; Shaw, R.M. Intracellular trafficking pathways of Cx43 gap junction channels. Biochim. Biophys. Acta BBA Biomembr. 2018, 1860, 40–47. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotini, A.; Martínez-Sarrà, E.; Pozzo, E.; Sampaolesi, M. Interactions between microRNAs and long non-coding RNAs in cardiac development and repair. Pharmacol. Res. 2018, 127, 58–66. [Google Scholar] [CrossRef]
- Kay, M.; Soltani, B.M.; Aghdaei, F.H.; Ansari, H.; Baharvand, H. Hsa-miR-335 regulates cardiac mesoderm and progenitor cell differentiation. Stem Cell Res. Ther. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Ekhteraei-Tousi, S.; Mohammad-Soltani, B.; Sadeghizadeh, M.; Mowla, S.J.; Parsi, S.; Soleimani, M. Inhibitory effect of hsa-miR-590-5p on cardiosphere-derived stem cells differentiation through downregulation of TGFB signaling. J. Cell. Biochem. 2015, 116, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, M.; Soltani, B.M. Long Noncoding RNA LOC400043 (LINC02381) Inhibits Gastric Cancer Progression Through Regulating Wnt Signaling Pathway. Front. Oncol. 2020, 10, 2189. [Google Scholar] [CrossRef]
- Melé, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlackow, M.; Nojima, T.; Gomes, T.; Dhir, A.; Carmo-Fonseca, M.; Proudfoot, N.J. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell 2017, 65, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Osak, M.; Bogu, G.K.; Stanton, L.W.; Johnson, R.; Lipovich, L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 2010, 16, 1478–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Washietl, S.; Kellis, M.; Garber, M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014, 24, 616–628. [Google Scholar] [CrossRef] [Green Version]
- Marques, A.C.; Ponting, C.P. Catalogues of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol. 2009, 10, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulitsky, I.; Shkumatava, A.; Jan, C.H.; Sive, H.; Bartel, D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011, 147, 1537–1550. [Google Scholar] [CrossRef] [Green Version]
- Kapusta, A.; Feschotte, C. Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef] [Green Version]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [Green Version]
- Rinn, J.L.; Chang, H.Y. Long noncoding RNAs: Molecular modalities to organismal functions. Annu. Rev. Biochem. 2020, 89, 283–308. [Google Scholar] [CrossRef]
- Laurent, G.S.; Wahlestedt, C.; Kapranov, P. The Landscape of long noncoding RNA classification. Trends Genet. 2015, 31, 239–251. [Google Scholar] [CrossRef] [Green Version]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, E.J.; Chin-Inmanu, K.; Jia, H.; Lipovich, L. Sense-antisense gene pairs: Sequence, transcription, and structure are not conserved between human and mouse. Front. Genet. 2013, 4, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yan, P.; et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 2016, 18, 637–652. [Google Scholar] [CrossRef]
- Zampetaki, A.; Albrecht, A.; Steinhofel, K. Long non-coding RNA structure and function: Is there a link? Front. Physiol. 2018, 9, 1201. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, R.B.-T.; Ulitsky, I. The functions of long noncoding RNAs in development and stem cells. Development 2016, 143, 3882–3894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjeije, H.; Soltani, B.M.; Behmanesh, M.; Zali, M.R. YWHAE long non-coding RNA competes with miR-323a-3p and miR-532-5p through activating K-Ras/Erk1/2 and PI3K/Akt signaling pathways in HCT116 cells. Hum. Mol. Genet. 2019, 28, 3219–3231. [Google Scholar] [CrossRef]
- Ounzain, S.; Pedrazzini, T. Super-enhancer lncs to cardiovascular development and disease. Biochim. Biophys. Acta BBA Mol. Cell Res. 2016, 1863, 1953–1960. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Han, P.; Li, W.; Lin, C.-H.; Yang, J.; Shang, C.; Nurnberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.-Y.; Lin, C.-J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnusamy, M.; Liu, F.; Zhang, Y.-H.; Li, R.-B.; Zhai, M.; Liu, F.; Zhou, L.-Y.; Liu, C.-Y.; Yan, K.-W.; Dong, Y.-H.; et al. Long noncoding RNA CPR (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation 2019, 139, 2668–2684. [Google Scholar] [CrossRef]
- Kang, X.; Zhao, Y.; Van Arsdell, G.; Nelson, S.F.; Touma, M. Ppp1r1b-lncRNA inhibits PRC2 at myogenic regulatory genes to promote cardiac and skeletal muscle development in mouse and human. RNA 2020, 26, 481–491. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016, 111, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiao, L.; Sun, L.; Li, Y.; Gao, Y.; Xu, C.; Shao, Y.; Li, M.; Li, C.; Lu, Y.; et al. LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca2+ overload and contractile dysfunction in a mouse model of myocardial infarction. Circ. Res. 2018, 122, 1354–1368. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Zhang, Y.; Zhao, Y.; Wang, J.; Li, T.; Zhang, Y.; Jiang, Y.; Jin, X.; Xue, G.; Li, P.; et al. Long Noncoding RNA–DACH1 (Dachshund Homolog 1) Regulates Cardiac Function by Inhibiting SERCA2a (Sarcoplasmic Reticulum Calcium ATPase 2a). Hypertension 2019, 74, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Chen, G.; Lv, F.; Liu, Y.; Tian, H.; Tao, R.; Jiang, R.; Zhang, W.; Zhuo, C. LncRNA TINCR attenuates cardiac hypertrophy by epigenetically silencing CaMKII. Oncotarget 2017, 8, 47565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Sun, L.; Xuan, L.; Pan, Z.; Hu, X.; Liu, H.; Bai, Y.; Jiao, L.; Li, Z.; Cui, L.; et al. Long non-coding RNA CCRR controls cardiac conduction via regulating intercellular coupling. Nat. Commun. 2018, 9, 4176. [Google Scholar] [CrossRef] [Green Version]
- Long, Q.Q.; Wang, H.; Gao, W.; Fan, Y.; Li, Y.F.; Ma, Y.; Yang, Y.; Shi, H.J.; Chen, B.R.; Meng, H.Y.; et al. Long noncoding RNA Kcna2 antisense RNA contributes to ventricular arrhythmias via silencing Kcna2 in rats with congestive heart failure. J. Am. Heart Assoc. 2017, 6, e005965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, P.; Yang, M.; Ren, H.; Shen, G.; Chen, J.; Zhang, J.; Liu, J.; Sun, C. Long noncoding RNA MALAT1 downregulates cardiac transient outward potassium current by regulating miR-200c/HMGB1 pathway. J. Cell. Biochem. 2018, 119, 10239–10249. [Google Scholar] [CrossRef]
- Dai, W.; Chao, X.; Li, S.; Zhou, S.; Zhong, G.; Jiang, Z. Long Noncoding RNA HOTAIR Functions as a Competitive Endogenous RNA to Regulate Connexin43 Remodeling in Atrial Fibrillation by Sponging MicroRNA-613. Cardiovasc. Ther. 2020, 2020, 5925342. [Google Scholar] [CrossRef]
- Wang, K.; Long, B.; Zhou, L.-Y.; Liu, F.; Zhou, Q.-Y.; Liu, C.-Y.; Fan, Y.-Y.; Li, P.-F. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat. Commun. 2014, 5, 3596. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Sun, T.; Li, N.; Wang, Y.; Wang, J.-X.; Zhou, L.-Y.; Long, B.; Liu, C.-Y.; Liu, F.; Li, P.-F. MDRL lncRNA regulates the processing of miR-484 primary transcript by targeting miR-361. PloS Genet. 2014, 10, e1004467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zou, X.; Liu, F. Silencing TTTY15 mitigates hypoxia-induced mitochondrial energy metabolism dysfunction and cardiomyocytes apoptosis via TTTY15/let-7i-5p and TLR3/NF-κB pathways. Cell Signal. 2020, 76, 109779. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Xu, Y.; Liang, C.; Xing, W.; Zhang, T. The mechanism of myocardial hypertrophy regulated by the interaction between mhrt and myocardin. Cell Signal. 2018, 43, 11–20. [Google Scholar] [CrossRef]
- Xu, Y.; Luo, Y.; Liang, C.; Zhang, T. LncRNA-Mhrt regulates cardiac hypertrophy by modulating the miR-145a-5p/KLF4/myocardin axis. J. Mol. Cell. Cardiol. 2020, 139, 47–61. [Google Scholar] [CrossRef]
- Viereck, J.; Bührke, A.; Foinquinos, A.; Chatterjee, S.; Kleeberger, J.A.; Xiao, K.; Janssen-Peters, H.; Batkai, S.; Ramanujam, D.; Kraft, T.; et al. Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur. Heart J. 2020, 41, 3462–3474. [Google Scholar]
- Li, X.; Wang, H.; Yao, B.; Xu, W.; Chen, J.; Zhou, X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci. Rep. 2016, 6, 36340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Dong, B.; Chen, M.; Yao, C. LncRNA H19 suppresses pyroptosis of cardiomyocytes to attenuate myocardial infarction in a PBX3/CYP1B1-dependent manner. Mol. Cell. Biochem. 2021, 476, 1387–1400. [Google Scholar] [CrossRef] [PubMed]
- Juan, V.; Crain, C.; Wilson, C. Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 2000, 28, 1221–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, L.; Li, M.; Shao, Y.; Zhang, Y.; Gong, M.; Yang, X.; Wang, Y.; Tan, Z.; Sun, L.; Xuan, L.; et al. lncRNA-ZFAS1 induces mitochondria-mediated apoptosis by causing cytosolic Ca 2+ overload in myocardial infarction mice model. Cell Death Dis. 2019, 10, 942. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ma, W.; Wang, X.; Sukhareva, N.; Hua, B.; Zhang, L.; Xu, J.; Li, X.; Li, S.; Liu, S.; et al. Targeting LncDACH1 promotes cardiac repair and regeneration after myocardium infarction. Cell Death Differ. 2020, 27, 2158–2175. [Google Scholar] [CrossRef]
- Trembinski, D.J.; Bink, D.I.; Theodorou, K.; Sommer, J.; Fischer, A.; van Bergen, A.; Kuo, C.-C.; Costa, I.G.; Schürmann, C.; Leisegang, M.S.; et al. Aging-regulated anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.-J.; Ji, Y.-X.; Zhang, P.; Deng, K.-Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016, 22, 1131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liang, Y.; Huang, X.; Guo, X.; Liu, Y.; Zhong, J.; Yuan, J. STAT3-induced upregulation of lncRNA MEG3 regulates the growth of cardiac hypertrophy through miR-361-5p/HDAC9 axis. Sci. Rep. 2019, 9, 460. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, J.; Geng, J.; Chen, F.; Wei, Z.; Liu, C.; Zhang, X.; Li, Q.; Zhang, J.; Gao, L.; et al. Long non-coding RNA MEG3 knockdown attenuates endoplasmic reticulum stress-mediated apoptosis by targeting p53 following myocardial infarction. J. Cell. Mol. Med. 2019, 23, 8369–8380. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Li, T.; Li, X.; Xu, C.; Liu, Q.; Jiang, H.; Li, Y.; Liu, Y.; Yan, H.; Huang, Q.; et al. The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR-214. Mol. Ther. Nucleic Acids 2018, 10, 387–397. [Google Scholar] [CrossRef] [Green Version]
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016, 8, 326ra22. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.-Y.; Long, B.; Yuan, S.-M.; Wang, Y.; Liu, C.-Y.; Sun, T.; Zhang, X.-J.; Li, P.-F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.M.; Li, H.; Shu, Q.; Wu, W.J.; Luo, X.M.; Lu, L. LncRNA SNHG1 exerts a protective role in cardiomyocytes hypertrophy via targeting miR-15a-5p/HMGA1 axis. Cell Biol. Int. 2020, 44, 1009–1019. [Google Scholar] [CrossRef]
- Zhang, N.; Meng, X.; Mei, L.; Hu, J.; Zhao, C.; Chen, W. The long non-coding RNA SNHG1 attenuates cell apoptosis by regulating miR-195 and BCL2-like protein 2 in human cardiomyocytes. Cell. Physiol. Biochem. 2018, 50, 1029–1040. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Zhang, Z.; Lu, X.; Yang, X. Long non-coding RNA SNHG15 regulates cardiomyocyte apoptosis after hypoxia/reperfusion injury via modulating miR-188-5p/PTEN axis. Arch. Physiol. Biochem. 2020, 1–8. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, W.; Xue, R.; Dong, B.; Liang, Z.; Chen, C.; Li, J.; Wang, Y.; Zhao, J.; Huang, H.; et al. Transcribed Ultraconserved Regions, Uc. 323, Ameliorates Cardiac Hypertrophy by Regulating the Transcription of CPT1b (Carnitine Palmitoyl transferase 1b). Hypertension 2020, 75, 79–90. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, F.; Wang, F.; Wu, N. Long noncoding RNA MAGI1-IT1 regulates cardiac hypertrophy by modulating miR-302e/DKK1/Wnt/beta-catenin signaling pathway. J. Cell. Physiol. 2020, 235, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, X.; Li, B.; Wang, H.; Li, M.; Huang, S.; Sun, Y.; Chen, G.; Si, X.; Huang, C. Long non-coding RNA ECRAR triggers post-natal myocardial regeneration by activating ERK1/2 signaling. Mol. Ther. 2019, 27, 29–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Li, H.; Li, X.; Li, B.; Zhong, L.; Huang, S.; Zheng, H.; Li, M.; Jin, G.; Liao, W.; et al. Loss of long non-coding RNA CRRL promotes cardiomyocyte regeneration and improves cardiac repair by functioning as a competing endogenous RNA. J. Mol. Cell. Cardiol. 2018, 122, 152–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Li, C.; Feng, J.; Zhang, J.; Fang, Y. lncRNA UCA1 is a novel regulator in cardiomyocyte hypertrophy through targeting the miR-184/HOXA9 axis. Cardiorenal Med. 2018, 8, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, X.; Shen, D.; Ge, D.; Chen, J.; Pei, J.; Li, Y.; Yue, Z.; Feng, J.; Chu, M.; et al. A long noncoding RNA NR_045363 controls cardiomyocyte proliferation and cardiac repair. J. Mol. Cell. Cardiol. 2019, 127, 105–114. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Cai, B.; Li, Y.; Jiang, Y.; Fu, X.; Zhao, Y.; Gao, H.; Yang, Y.; Yang, J.; et al. The long noncoding RNA lncCIRBIL disrupts the nuclear translocation of Bclaf1 alleviating cardiac ischemia–reperfusion injury. Nat. Commun. 2021, 12, 522. [Google Scholar]
- Hang, C.T.; Yang, J.; Han, P.; Cheng, H.-L.; Shang, C.; Ashley, E.; Zhou, B.; Chang, C.-P. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010, 466, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lu, M.M.; Cheng, L.; Yuan, L.-J.; Zhu, X.; Stout, A.L.; Chen, M.; Li, J.; Parmacek, M.S. Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc. Natl. Acad. Sci. USA 2009, 106, 18734–18739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swaminathan, P.D.; Purohit, A.; Hund, T.J.; Anderson, M.E. Calmodulin-dependent protein kinase II: Linking heart failure and arrhythmias. Circ. Res. 2012, 110, 1661–1677. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Xu, H.; Cheng, J.; Zhang, Y.; Gao, C.; Fan, T.; Peng, B.; Li, B.; Liu, L.; Cheng, Z. Downregulation of long non-coding RNA H19 promotes P19CL6 cells proliferation and inhibits apoptosis during late-stage cardiac differentiation via miR-19b-modulated Sox6. Cell Biosci. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Yang, Y.; Xu, Z.; Lan, C.; Chen, C.; Li, C.; Chen, Z.; Yu, C.; Xia, X.; Liao, Q.; et al. Long Noncoding RNA Ahit Protects Against Cardiac Hypertrophy Through SUZ12 (Suppressor of Zeste 12 Protein Homolog)-Mediated Downregulation of MEF2A (Myocyte Enhancer Factor 2A). Circ. Heart Fail. 2020, 13, e006525. [Google Scholar] [CrossRef]
- Pereira, A.H.M.; Cardoso, A.C.; Consonni, S.R.; Oliveira, R.R.; Saito, A.; Vaggione, M.L.B.; Matos-Souza, J.R.; Carazzolle, M.F.; Goncalves, A.; Fernandes, J.L. MEF2C repressor variant deregulation leads to cell cycle re-entry and development of heart failure. EBioMedicine 2020, 51, 102571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Oort, R.; van Rooij, E.; Bourajjaj, M.; Schimmel, J.; Jansen, M.; van der Nagel, R.; Doevendans, P.; Schneider, M.; van Echteld, C.; De Windt, L.J. MEF2 activates a genetic program promoting chamber dilation and contractile dysfunction in calcineurin-induced heart failure. Circulation 2006, 114, 298–308. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Ni, J.; Lan, L.; Michael, S.; Rui, G.; Xiyun, B.; Xiaozhi, L.; Guanwei, F. SERCA2a: A key protein in the Ca2+ cycle of the heart failure. Heart Fail. Rev. 2019, 25, 523–535. [Google Scholar]
- Tian, Y.; Liu, Y.; Wang, T.; Zhou, N.; Kong, J.; Chen, L.; Snitow, M.; Morley, M.; Li, D.; Petrenko, N.; et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med. 2015, 7, 279ra38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohjoismäki, J.L.; Goffart, S. The role of mitochondria in cardiac development and protection. Free Radic. Biol. Med. 2017, 106, 345–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, D.; Jian, C.; Peng, Q.; Hou, T.; Wu, K.; Shang, B.; Zhao, M.; Wang, Y.; Zheng, W.; Ma, Q.; et al. Prohibitin 2 deficiency impairs cardiac fatty acid oxidation and causes heart failure. Cell Death Dis. 2020, 11, 181. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Long, B.; Jiao, J.-Q.; Wang, J.-X.; Liu, J.-P.; Li, Q.; Li, P.-F. miR-484 regulates mitochondrial network through targeting Fis1. Nat. Commun. 2012, 3, 781. [Google Scholar] [CrossRef]
- Forsburg, S.L. Eukaryotic MCM proteins: Beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004, 68, 109–131. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Tan, B.; Yang, Z.; Yu, X.; Chen, L.; Ran, D.; Xu, Q.; Zhou, X. Nrf2/ARE pathway activation is involved in negatively regulating heat-induced apoptosis in non-small cell lung cancer cells. Acta Biochim. Biophys. Sin. 2020, 52, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Jaé, N.; Holdt, L.; Dimmeler, S. Long noncoding RNAs: From clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar] [CrossRef]
- Choong, O.K.; Lee, D.S.; Chen, C.-Y.; Hsieh, P.C. The roles of non-coding RNAs in cardiac regenerative medicine. Non-Coding RNA Res. 2017, 2, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Sarropoulos, I.; Marin, R.; Cardoso-Moreira, M.; Kaessmann, H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 2019, 571, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.J.; van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primates. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Busch, A.; Jin, H.; Chernogubova, E.; Pelisek, J.; Karlsson, J.; Sennblad, B.; Liu, S.; Lao, S.; Hofmann, P.; et al. H19 induces abdominal aortic aneurysm development and progression. Circulation 2018, 138, 1551–1568. [Google Scholar] [CrossRef] [PubMed]


| Maturation Characteristics | LncRNA | Function | Regulated Target | Ref |
|---|---|---|---|---|
| Myofibril formation | Mhrt | Regulation of Myh6/Myh7 ratio | Brg1 | [77] |
| Cpr | Sarcomere organization | Mcm3 | [78] | |
| Ppp1r1b | Cardiac myogenesis regulation | Tcap | [79] | |
| Electrophysiology | H19 | CaMKIIδ expression regulation | miR-675 | [80] |
| Zfas1 | Ca2+ homeostasis regulation | Serca2a | [81] | |
| Dach1 | Ca2+ homeostasis regulation | Serca2a | [82] | |
| Tincr | CaMKII expression regulation | EZH2 | [83] | |
| Ccrr | Cardiac conduction regulation | CIP85 | [84] | |
| Kcna2-AS | Cardiac conduction regulation | Kv1.2 | [85] | |
| Malat1 | Cardiac conduction regulation | miR-200c | [86] | |
| ZNF593-AS | Cardiac conduction regulation | HNRNPC | ||
| Hotair | Regulate Cx43 expression | miR-613 | [87] | |
| Metabolism | Carl | Regulate metabolic maturation | miR-539 | [88] |
| Mdrl | Regulate metabolic maturation | miR-361miR-484 | [89] | |
| TTTY15 | Regulate mitochondrial energy metabolism | let-7i-5p | [90] | |
| Proliferation and hypertrophy | Mhrt | Hypertrophy regulation | Hdac5 | [91] |
| Hypertrophy regulation/Myocd regulation | miR-145-5p | [92] | ||
| H19 | Hypertrophy regulation/NFAT signaling | Prc2 | [93] | |
| Mitochondria-mediated apoptosis regulation by PBX3-dependent way | miR-675 CYP1B1 | [94] | ||
| [95] | ||||
| Ahit | Hypertrophy regulation/Mef2A regulation | Suz12-Prc2 | [96] | |
| Zfas1 | Mitochondria-mediated apoptosis | Serca2a | [97] | |
| Dach1 | Proliferation regulation/Hippo signaling | PP1A | [98] | |
| Cpr | Proliferation regulation | Mcm3 | [78] | |
| Sarrah | Apoptosis and survival regulation/Nrf2 pathway | CRIP2 and p300 | [99] | |
| Chaer | Hypertrophy regulation | Prc2 | [100] | |
| Meg3 | Hypertrophy regulation | miR-361-5p | [101] | |
| Apoptosis regulation/NF-κB signaling | P53 | [102] | ||
| Plscr4 | Hypertrophy regulation | miR-214 | [103] | |
| Chast | Hypertrophy regulation | Plekhm1 | [104] | |
| Chrf | Hypertrophy regulation | miR-489 | [105] | |
| Snhg1 | Hypertrophy regulation | miR-15a-5p | [106] | |
| Apoptosis regulation | miR-195 | [107] | ||
| Apoptosis regulation | miR-188-5p | [108] | ||
| Uc.323 | Hypertrophy regulation | Ezh2 | [109] | |
| Magi1-IT1 | Hypertrophy regulation/Wnt signaling | miR-302e | [110] | |
| Ecrar | Proliferation regulation/ERK1/2 signaling | ERK1/2 | [111] | |
| Crrl | Proliferation regulation/HOPX regulation | miR-199a-3p | [112] | |
| Uca1 | Hypertrophy regulation | miR-184 | [113] | |
| NR_045363 | Proliferation regulation/JAK2-STAT3 signaling | miR-216a | [114] | |
| lncCIRBIL | Apoptosis regulation | Bclaf1 | [115] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kay, M.; Soltani, B.M. LncRNAs in Cardiomyocyte Maturation: New Window for Cardiac Regenerative Medicine. Non-Coding RNA 2021, 7, 20. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010020
Kay M, Soltani BM. LncRNAs in Cardiomyocyte Maturation: New Window for Cardiac Regenerative Medicine. Non-Coding RNA. 2021; 7(1):20. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010020
Chicago/Turabian StyleKay, Maryam, and Bahram M. Soltani. 2021. "LncRNAs in Cardiomyocyte Maturation: New Window for Cardiac Regenerative Medicine" Non-Coding RNA 7, no. 1: 20. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010020




