Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message
Abstract
:1. Introduction
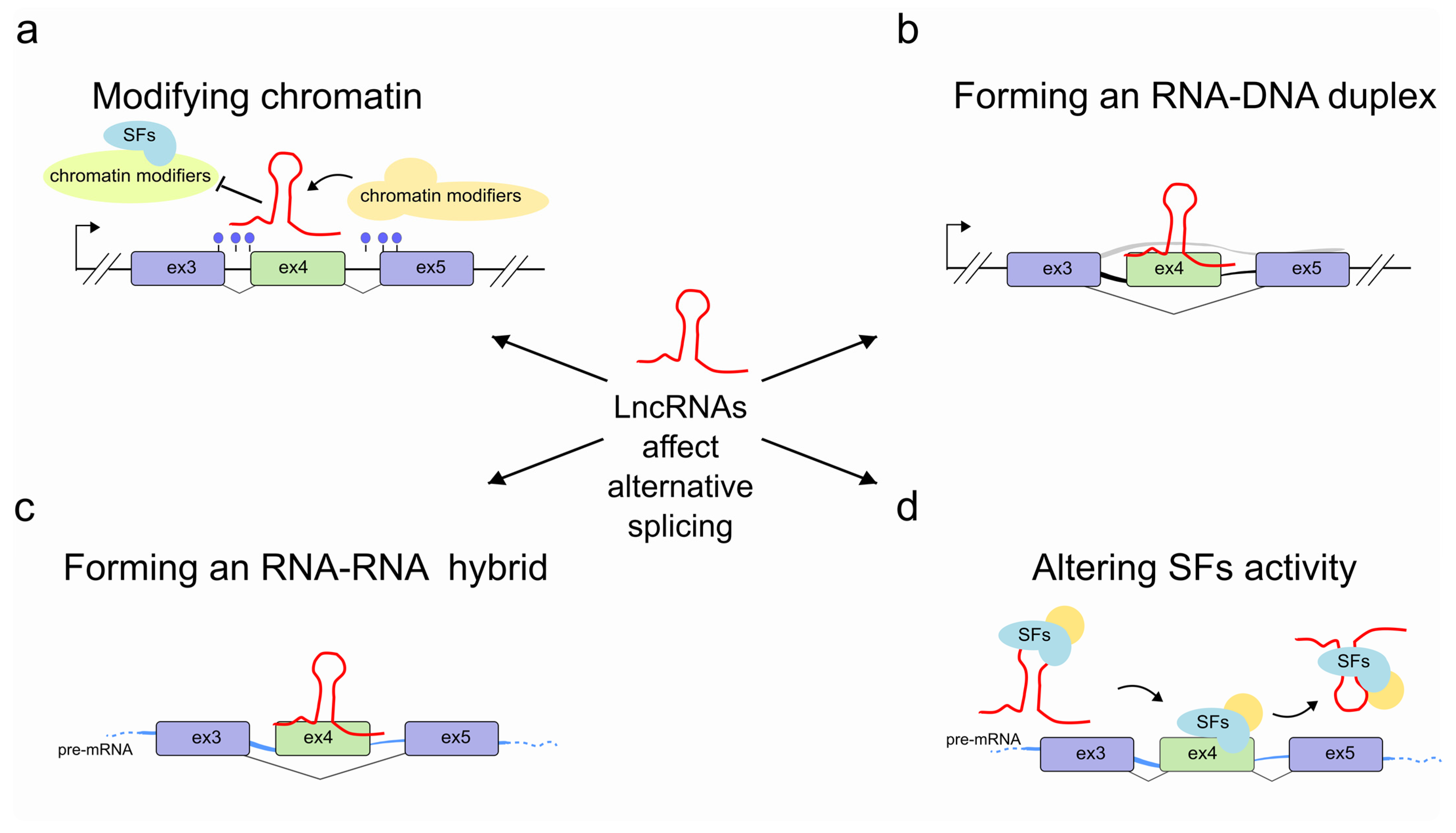
2. LncRNAs Regulate Alternative Splicing through Chromatin Modification
3. LncRNAs Regulate Pre-mRNA Splicing through RNA-DNA Interactions
4. LncRNAs Regulate Pre-mRNA Splicing through RNA-RNA Interactions
5. LncRNAs Regulate Pre-mRNA Splicing by Modulating the Activity of Splicing Factors
6. Concluding Remarks and Future Perspectives
| LncRNA Name | Splicing Target | Splicing Mechanism | Regulatory Effect or Associated Disease | Ref |
|---|---|---|---|---|
| LncRNAs regulating AS by chromatin modifications | ||||
| asFGFR2 | FGFR2 | Recruiting Polycomb complexes and KDM2a to modify histone methylation and favor exon IIIb inclusion | Epithelial development | [23] |
| Antisense transcripts at each Pcdhα first exon | Pcdhα | First exon selection by histone modifications and distant DNA loop | Neuronal self-identity | [31] |
| LncRNAs regulate AS through DNA-RNA interactions | ||||
| SEP3 exon6 circRNA (plant) | SEP3 | Exon skipping through R-loop formation at exon 6 | Flowering time | [44] |
| LncRNAs regulate AS through RNA-RNA interactions | ||||
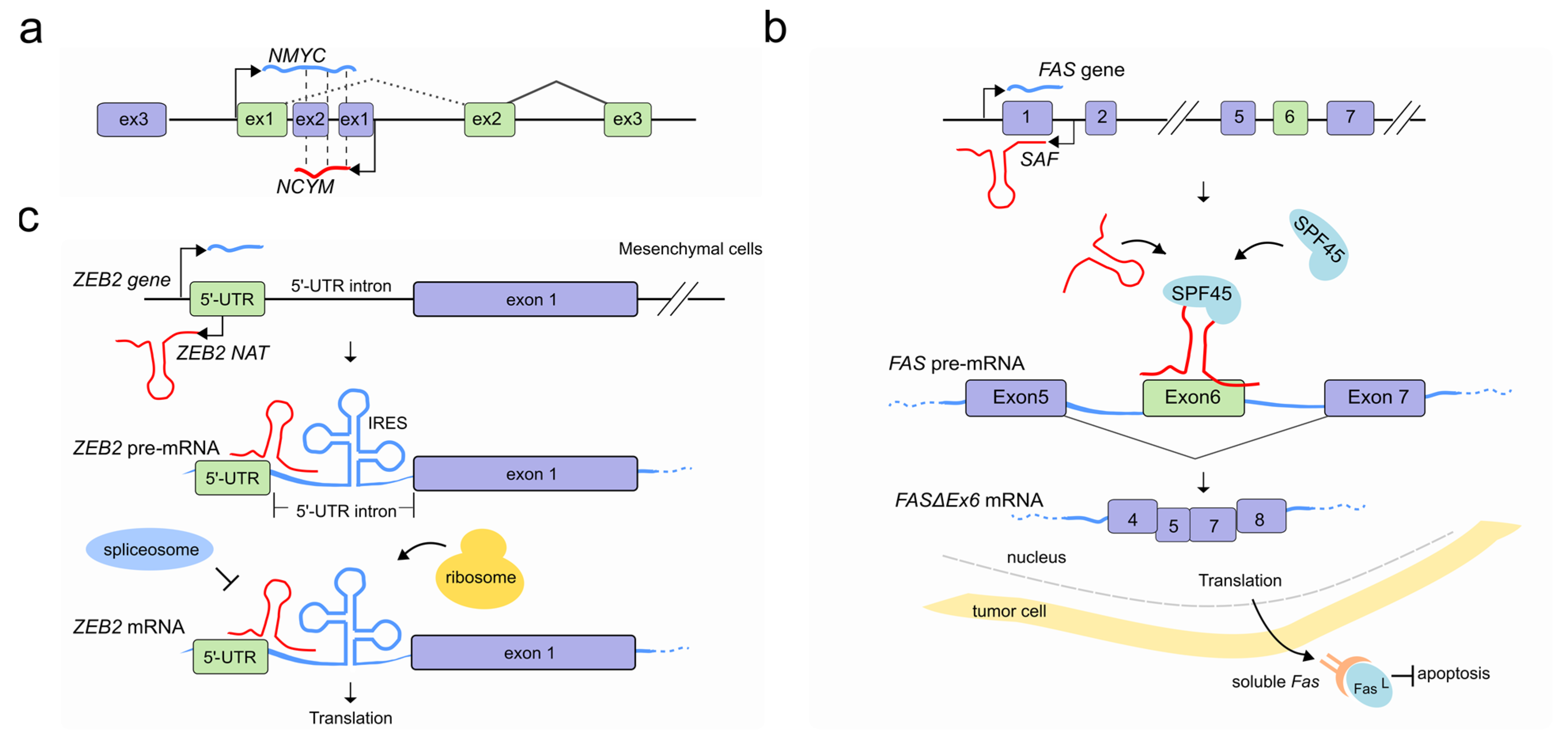
| NCYM NAT | NMYC | Intron I retention via antisense-sense RNA-RNA duplex | Cancer | [57] |
| NR1D1 | THRA | Favoring α1 isoform by forming antisense-sense RNA-RNA duplex with the α2 mRNA | Thyroid hormone-responsiveness | [58,59] |
| SAF | FAS | Exon 6 skipping by forming RNA-RNA duplex with the target pre-mRNA and recruiting SPF45 | Cancer Apoptosis | [60] |
| ZEB2 NAT | ZEB2 | Preventing splicing of the IRES-containing intron through RNA-RNA interaction with the mRNA | EMT | [61] |
| 51A | SORL1 | Splicing shift from A to variant B by antisense-sense RNA-RNA duplex with an intronic sequence of the pre-mRNA | Alzheimer | [67] |
| 17A | GPR51 | Splicing shift from full-length to shorter GABAB R2 variant by antisense-sense RNA-RNA duplex | Alzheimer | [68] |
| LncRNAs regulate AS by modulating the activity of splicing factors | ||||
| MALAT1/NEAT2 | Modulation of SR localization and phosphorylation Uncoupling PTBP2 from SFPQ-PTBP2 | Cancer | [73,88] | |
| NEAT1 | PPARγ | By interacting with CLK1 kinase to modulate SRp40 phosphorylation status | Adipocyte differentiation | [71,89,90] |
| Gomafu/RNCR2/ MIAT | Interaction with QKI and SRSF1 Association with SF1 Localization of SF1 and Clf3 in CS bodies | Schizophrenia Retinal cell and brain development Post-mitotic neuronal function | [97,98,99,100] | |
| LINC01133 | Interaction and titration of SRSF6 splicing factor from target genes | EMT | [101] | |
| PNCTR | Hijacking PTBP1 in the perinucleolar compartment | Cell survival | [102] | |
| TPM1-AS | TPM1 | Splicing shift to V1 or V3 isoforms by sequestering RBM4 | Cancer | [107] |
| ASCO (plant) | Association with SmD1b and PRP8a and hijacking NSRa/b from the spliceosome | Lateral root formation | [109,110] | |
| ENOD40 (plant) | Control nucleocytoplasmic of MtRBP1 | Symbiotic nodule development | [111,112] | |
| PCGEM1 | Mutual bond with either hnRNP A1 or U2AF65 to promote or suppress specific AR splice variants | Castration resistance | [113] | |
| BC200 | BCL-x | Interaction with pre-mRNA and recruitment of the hnRNP A2/B1 which prevent Sam68 association | Apoptosis | [115,117] |
| Lnc-Spry1 | Interaction with U2AF65 | EMT | [118] | |
| LASTR | Promoting splicing efficiency by interacting with SART3 | Stress-induced JNK/c-JUN pathway | [119] | |
| LINC-HELLP | Interaction with ribosomal and splicing complex components (eg: YBX1, PCBP1, PCBP2, RPS6 and RPL7) | HELLP syndrome | [120] | |
| DSCAM-AS1 | Exon skipping and 3′ UTR usage by interaction with hnRNPL | Tumor progression and anti-estrogen resistance | [121] | |
| CircMbl | Mbl | Competing with the linear cognate by sequestering Mbl protein | Neuron Development | [123] |
| CircSMARCA5 | Interaction with SRSF1 and promotion of the anti-angiogenic splicing isoforms of VEGF-A | Angiogenesis | [124] | |
| PNUTS | PNUTS | Self-splicing regulation modulating the activity of hnRNP E1 | EMT | [126] |
Funding
Conflicts of Interest
Abbreviations
References
- Berget, S.M.; Moore, C.; Sharp, P.A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. USA 1977, 74, 3171–3175. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655–670. [Google Scholar] [CrossRef]
- Rosenfeld, M.G.; Amara, S.G.; Roos, B.A.; Ong, E.S.; Evans, R.M. Altered expression of the calcitonin gene associated with RNA polymorphism. Nature 1981, 290, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Ward, A.J.; Cooper, T.A. The pathobiology of splicing. J. Pathol. 2010, 220, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Magen, A.; Ast, G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007, 35, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattick, J.S. RNA regulation: A new genetics? Nat. Rev. Genet. 2004, 5, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- David Wang, X.Q.; Crutchley, J.L.; Dostie, J. Shaping the Genome with Non-coding RNAs. Curr. Genom. 2011, 307–321. [Google Scholar] [CrossRef]
- Flynn, R.A.; Chang, H.Y. Active chromatin and noncoding RNAs: An intimate relationship. Curr. Opin. Genet. Dev. 2012, 22, 172–178. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [Green Version]
- Goff, L.A.; Rinn, J.L. Linking RNA biology to lncRNAs. Genome Res. 2015, 25, 1456–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [Green Version]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef] [Green Version]
- Schor, I.E.; Rascovan, N.; Pelisch, F.; Alió, M.; Kornblihtt, A.R. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 4325–4330. [Google Scholar] [CrossRef] [Green Version]
- Batsché, E.; Yaniv, M.; Muchardt, C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct Mol. Biol. 2006. [Google Scholar] [CrossRef] [PubMed]
- Luco, R.F.; Allo, M.; Schor, I.E.; Kornblihtt, A.R.; Misteli, T. Epigenetics in alternative pre-mRNA splicing. Cell 2011, 144, 16–26. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, I.; Munita, R.; Agirre, E.; Dittmer, T.A.; Gysling, K.; Misteli, T.; Luco, R.F. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat. Struct. Mol. Biol. 2015, 22, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Du, J.; Sun, B.; Dong, X.; Xu, G.; Zhou, J.; Huang, Q.; Liu, Q.; Hao, Q.; Ding, J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006, 34, 6621–6628. [Google Scholar] [CrossRef] [PubMed]
- Kornblihtt, A.R. Chromatin, transcript elongation and alternative splicing. Nat. Struct. Mol. Biol. 2006, 13, 5–7. [Google Scholar] [CrossRef]
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, 74–79. [Google Scholar] [CrossRef]
- Amaral, P.P.; Leonardi, T.; Han, N.; Viré, E.; Gascoigne, D.K.; Arias-Carrasco, R.; Büscher, M.; Pandolfini, L.; Zhang, A.; Pluchino, S.; et al. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 2018, 19, 32. [Google Scholar] [CrossRef]
- Pisignano, G.; Pavlaki, I.; Murrell, A. Being in a loop: How long non-coding RNAs organise genome architecture. Essays Biochem. 2019, 63, 177–186. [Google Scholar] [CrossRef]
- Lefevre, P.; Witham, J.; Lacroix, C.E.; Cockerill, P.N.; Bonifer, C. The LPS-Induced Transcriptional Upregulation of the Chicken Lysozyme Locus Involves CTCF Eviction and Noncoding RNA Transcription. Mol. Cell. 2008, 32, 129–139. [Google Scholar] [CrossRef]
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019, 26, 2904–2915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canzio, D.; Nwakeze, C.L.; Horta, A.; Rajkumar, S.M.; Coffey, E.L.; Duffy, E.E.; Duffié, R.; Monahan, K.; O’Keeffe, S.; Simon, M.D.; et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin α Promoter Choice. Cell 2019, 177, 639–653. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Monahan, K.; Wu, H.; Gertz, J.; Varley, K.E.; Li, W.; Myers, R.M.; Maniatis, T.; Wu, Q. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl. Acad. Sci. USA 2012, 109, 21081–21086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehayova, P.; Monahan, K.; Chen, W.; Maniatis, T. Regulatory elements required for the activation and repression of the protocadherin-á gene cluster. Proc. Natl. Acad. Sci. USA 2011, 108, 17195–17200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monahan, K.; Rudnick, N.D.; Kehayova, P.D.; Pauli, F.; Newberry, K.M.; Myers, R.M.; Maniatis, T. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of Protocadherin-α gene expression. Proc. Natl. Acad. Sci. USA 2012, 109, 9125–9130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribich, S.; Tasic, B.; Maniatis, T. Identification of long-range regulatory elements in the protocadherin-α gene cluster. Proc. Natl. Acad. Sci. USA 2006, 103, 19719–19724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, S.P.; Wang, P.L.; Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015, 4, e07540. [Google Scholar] [CrossRef]
- Lee, E.C.S.; Elhassan, S.A.M.; Lim, G.P.L.; Kok, W.H.; Tan, S.W.; Leong, E.N.; Tan, S.H.; Chan, E.W.L.; Bhattamisra, S.K.; Rajendran, R.; et al. The roles of circular RNAs in human development and diseases. Biomed. Pharmacother. 2019, 111, 198–208. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang HBin Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Alexander, R.D.; Innocente, S.A.; Barrass, J.D.; Beggs, J.D. Splicing-Dependent RNA polymerase pausing in yeast. Mol. Cell. 2010, 40, 582–593. [Google Scholar] [CrossRef] [PubMed]
- El Hage, A.; Webb, S.; Kerr, A.; Tollervey, D. Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria. PLoS Genet. 2014, 10, e1004716. [Google Scholar] [CrossRef] [PubMed]
- Wongsurawat, T.; Jenjaroenpun, P.; Kwoh, C.K.; Kuznetsov, V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugino, A.; Hirose, S.; Okazaki, R. RNA-linked nascent DNA fragments in Escherichia coli. Proc. Natl. Acad. Sci. USA 1972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greider, C.W.; Blackburn, E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 1989. [Google Scholar] [CrossRef]
- Williams, J.S.; Kunkel, T.A. Ribonucleotides in DNA: Origins, repair and consequences. DNA Repair (Amst.) 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013. [Google Scholar] [CrossRef] [PubMed]
- Morrissy, A.S.; Griffith, M.; Marra, M.A. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aartsma-Rus, A.; Van Ommen, G.J.B. Antisense-mediated exon skipping: A versatile tool with therapeutic and research applications. RNA 2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khorkova, O.; Myers, A.J.; Hsiao, J.; Wahlestedt, C. Natural antisense transcripts. Hum. Mol. Genet. 2014. [Google Scholar] [CrossRef] [Green Version]
- Bardou, F.; Merchan, F.; Ariel, F.; Crespi, M. Dual RNAs in plants. Biochimie 2011. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, Y.; Islam, S.M.; Alagu, J.; Kaneko, Y.; Kato, M.; Tanaka, Y.; Kawana, H.; Hossain, S.; Matsumoto, D.; Yamamoto, M.; et al. NCYM, a Cis-Antisense Gene of MYCN, Encodes a De Novo Evolved Protein That Inhibits GSK3β Resulting in the Stabilization of MYCN in Human Neuroblastomas. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef]
- Krystal, G.W.; Armstrong, B.C.; Battey, J.F. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol. Cell Biol. 1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munroe, S.H.; Lazar, M.A. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J. Biol Chem. 1991, 266, 22083–22086. [Google Scholar] [CrossRef]
- Chassande, O.; Fraichard, A.; Gauthier, K.; Flamant, F.; Legrand, C.; Savatier, P.; Laudet, V.; Samarut, J. Identification of Transcripts Initiated from an Internal Promoter in the c-erbAα Locus That Encode Inhibitors of Retinoic Acid Receptor-α and Triiodothyronine Receptor Activities. Mol. Endocrinol. 1997, 11, 1278–1290. [Google Scholar] [CrossRef] [Green Version]
- Villamizar, O.; Chambers, C.B.; Riberdy, J.M.; Persons, D.A.; Wilber, A. Long noncoding RNA Saf and splicing factor 45 increase soluble Fas and resistance to apoptosis. Oncotarget 2016. [Google Scholar] [CrossRef]
- Beltran, M.; Puig, I.; Peña, C.; García, J.M.; Alvarez, A.B.; Peña, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.L.; Galasko, D.R.; Ringman, J.M.; Vinters, H.V.; Edland, S.D.; Pomakian, J.; Ubeda, O.J.; Rosario, E.R.; Teter, B.; Frautschy, S.A.; et al. Reduction of SorLA/LR11, a sorting protein limiting β-amyloid production, in alzheimer disease cerebrospinal fluid. Arch. Neurol. 2009, 66, 448–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reitz, C.; Cheng, R.; Rogaeva, E.; Lee, J.H.; Tokuhiro, S.; Zou, F.; Bettens, K.; Sleegers, K.; Tan, E.K.; Kimura, R.; et al. Meta-analysis of the association between variants in SORL1 and Alzheimer disease. Arch. Neurol. 2011, 68, 99–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogaeva, E.; Meng, Y.; Lee, J.H.; Gu, Y.; Kawarai, T.; Zou, F.; Katayama, T.; Baldwin, C.T.; Cheng, R.; Hasegawa, H.; et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007, 39, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.M.; Reiche, J.; Schmidt, V.; Gotthardt, M.; Spoelgen, R.; Behlke, J.; von Arnim, C.A.; Breiderhoff, T.; Jansen, P.; Wu, X.; et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA 2005, 102, 13461–13466. [Google Scholar] [CrossRef] [Green Version]
- Small, S.A.; Kent, K.; Pierce, A.; Leung, C.; Kang, M.S.; Okada, H.; Honig, L.; Vonsattel, J.P.; Kim, T.W. Model-guided microarray implicates the retromer complex in Alzheimer’s disease. Ann. Neurol. 2005, 58, 909–919. [Google Scholar] [CrossRef]
- Ciarlo, E.; Massone, S.; Penna, I.; Nizzari, M.; Gigoni, A.; Dieci, G.; Russo, C.; Florio, T.; Cancedda, R.; Pagano, A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. DMM Dis Model. Mech. 2013, 6, 424–433. [Google Scholar] [CrossRef] [Green Version]
- Massone, S.; Vassallo, I.; Fiorino, G.; Castelnuovo, M.; Barbieri, F.; Borghi, R.; Tabaton, M.; Robello, M.; Gatta, E.; Russo, C.; et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011, 41, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Onodera, C.S.; Underwood, J.G.; Katzman, S.; Jacobs, F.; Greenberg, D.; Salama, S.R.; Haussler, D. Gene isoform specificity through enhancer-associated antisense transcription. PLoS ONE 2012, 7, e43511. [Google Scholar] [CrossRef] [Green Version]
- Stork, M.; Di Lorenzo, M.; Welch, T.J.; Crosa, J.H. Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J. Bacteriol. 2007. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patton, J.G.; Porro, E.B.; Galceran, J.; Tempst, P.; Nadal-Ginard, B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993. [Google Scholar] [CrossRef] [Green Version]
- Gozani, O.; Patton, J.G.; Reed, R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994. [Google Scholar] [CrossRef]
- Misteli, T.; Cáceres, J.F.; Clement, J.Q.; Krainer, A.R.; Wilkinson, M.F.; Spector, D.L. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Jamison, S.F.; Garcia-Blanco, M.A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 1997, 3, 1456–1467. [Google Scholar] [PubMed]
- Sanford, J.R.; Ellis, J.D.; Cazalla, D.; Cáceres, J.F. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15042–15047. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yario, T.A.; Steitz, J.A. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA 2004. [Google Scholar] [CrossRef] [Green Version]
- Stamm, S. Regulation of alternative splicing by reversible protein phosphorylation. J. Biol Chem. 2008. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Manley, J.L. A Complex Signaling Pathway Regulates SRp38 Phosphorylation and Pre-mRNA Splicing in Response to Heat Shock. Mol. Cell 2007. [Google Scholar] [CrossRef]
- Zhong, X.Y.; Ding, J.H.; Adams, J.A.; Ghosh, G.; Fu, X.D. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009. [Google Scholar] [CrossRef] [Green Version]
- Engreitz, J.M.; Sirokman, K.; McDonel, P.; Shishkin, A.A.; Surka, C.; Russell, P.; Grossman, S.R.; Chow, A.Y.; Guttman, M.; Lander, E.S. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell. 2014, 159, 188–199. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell. 2016, 165, 1267–1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Cao, C.; Ji, L.; Ye, R.; Wang, D.; Xia, C.; Wang, S.; Du, Z.; Hu, N.; Yu, X.; et al. RIC-seq for global in situ profiling of RNA–RNA spatial interactions. Nature 2020, 582, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Malakar, P.; Shilo, A.; Mogilevsky, A.; Stein, I.; Pikarsky, E.; Nevo, Y.; Benyamini, H.; Elgavish, S.; Zong, X.; Prasanth, K.V.; et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017, 77, 155–1167. [Google Scholar] [CrossRef] [Green Version]
- Ji, Q.; Zhang, L.; Liu, X.; Zhou, L.; Wang, W.; Han, Z.; Sui, H.; Tang, Y.; Wang, Y.; Liu, N.; et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J. Cancer. 2014, 111, 736–748. [Google Scholar] [CrossRef] [Green Version]
- Jiang, K.; Patel, N.A.; Watson, J.E.; Apostolatos, H.; Kleiman, E.; Hanson, O.; Hagiwara, M.; Cooper, D.R. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCβJII messenger ribonucleic acid. Endocrinology 2009, 150, 2087–2097. [Google Scholar] [CrossRef]
- Cooper, D.R.; Carter, G.; Li, P.; Patel, R.; Watson, J.E.; Patel, N.A. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARγ2 splicing during adipogenesis in 3T3-L1 cells. Genes 2014, 5, 1050–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackshaw, S.; Harpavat, S.; Trimarchi, J.; Cai, L.; Huang, H.; Kuo, W.P.; Weber, G.; Lee, K.; Fraioli, R.E.; Cho, S.H.; et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004, 2. [Google Scholar] [CrossRef] [PubMed]
- Sone, M.; Hayashi, T.; Tarui, H.; Agata, K.; Takeichi, M.; Nakagawa, S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007, 120, 2498–2506. [Google Scholar] [CrossRef] [Green Version]
- Rapicavoli, N.A.; Blackshaw, S. New meaning in the message: Noncoding RNAs and their role in retinal development. Dev. Dyn. 2009. [Google Scholar] [CrossRef]
- Rapicavoli, N.A.; Poth, E.M.; Blackshaw SRapicavoli, N.A.; Poth, E.M.; Blackshaw, S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev. Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.; Dinger, M.E.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008. [Google Scholar] [CrossRef] [Green Version]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [Green Version]
- Tsuiji, H.; Yoshimoto, R.; Hasegawa, Y.; Furuno, M.; Yoshida, M.; Nakagawa, S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 2011, 16, 479–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladd, A.N. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Mol. Cell Neurosci. 2013, 56, 456–464. [Google Scholar] [CrossRef] [Green Version]
- Ishizuka, A.; Hasegawa, Y.; Ishida, K.; Yanaka, K.; Nakagawa, S. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes Cells. 2014, 19, 704–721. [Google Scholar] [CrossRef] [Green Version]
- Kong, J.; Sun, W.; Li, C.; Wan, L.; Wang, S.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. Long non-coding RNA LINC01133 inhibits epithelial–mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016, 380, 476–484. [Google Scholar] [CrossRef]
- Yap, K.; Mukhina, S.; Zhang, G.; Tan, J.S.C.; Ong, H.S.; Makeyev, E.V. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 2018, 72, 525–540. [Google Scholar] [CrossRef]
- Bushell, M.; Stoneley, M.; Kong, Y.W.; Hamilton, T.L.; Spriggs, K.A.; Dobbyn, H.C.; Qin, X.; Sarnow, P.; Willis, A.E. Polypyrimidine Tract Binding Protein Regulates IRES-Mediated Gene Expression during Apoptosis. Mol. Cell 2006, 23, 401–412. [Google Scholar] [CrossRef]
- Bielli, P.; Bordi, M.; Di Biasio, V.; Sette, C. Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5′ splice site selection. Nucleic Acids Res. 2014, 42, 12070–12081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Bahi, N.; Llovera, M.; Comella, J.X.; Sanchis, D. Polypyrimidine tract binding proteins (PTB) regulate the expression of apoptotic genes and susceptibility to caspase-dependent apoptosis in differentiating cardiomyocytes. Cell Death Differ. 2009, 16, 1460–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porto, F.W.; Daulatabad, S.V.; Janga, S.C. Long non-coding RNA expression levels modulate cell-type-specific splicing patterns by altering their interaction landscape with RNA-binding proteins. Genes 2019, 10, 593. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.W.; Zhang, Y.L.; Liao LDi Li, E.M.; Xu, L.Y. Natural antisense transcript TPM1-AS regulates the alternative splicing of tropomyosin I through an interaction with RNA-binding motif protein 4. Int. J. Biochem. Cell Biol. 2017, 90, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.; Crespi, M. Long Noncoding RNA Modulates Alternative Splicing Regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Tran, V.D.T.; Souiai, O.; Romero-Barrios, N.; Crespi, M.; Gautheret, D. Detection of generic differential RNA processing events from RNA-seq data. RNA Biol. 2016, 13, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Rigo, R.; Bazin, J.; Romero-Barrios, N.; Moison, M.; Lucero, L.; Christ, A.; Benhamed, M.; Blein, T.; Huguet, S.; Charon, C.; et al. The Arabidopsis lncRNA ASCO modulates the transcriptome through interaction with splicing factors. EMBO Rep. 2020, 21, e48977. [Google Scholar] [CrossRef]
- Crespi, M.D.; Jurkevitch, E.; Poiret, M.; d’Aubenton-Carafa, Y.; Petrovics, G.; Kondorosi, E.; Kondorosi, A. Enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994, 13, 5099–5112. [Google Scholar] [CrossRef]
- Campalans, A.; Kondorosi, A.; Crespi, M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant. Cell 2004, 16, 1047–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhou, N.; Huang, J.; Ho, T.T.; Zhu, Z.; Qiu, Z.; Zhou, X.; Bai, C.; Wu, F.; Xu, M.; et al. Regulation of androgen receptor splice variant AR3 by PCGEM1. Oncotarget 2016, 7, 15481–15491. [Google Scholar] [CrossRef] [PubMed]
- Hetz, C. BCL-2 protein family. Essential regulators of cell death. Preface. Adv. Exp. Med. Biol. 2010. [Google Scholar]
- Minnt, A.J.; Boise, L.H.; Thompson, C.B. Bcl-XS Antagonizes the protective effects of Bcl-xL. J. Biol. Chem. 1996, 271, 6306–6312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paronetto, M.P.; Achsel, T.; Massiello, A.; Chalfant, C.E.; Sette, C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 2007, 176, 929–939. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Gupta, S.C.; Peng, W.X.; Zhou, N.; Pochampally, R.; Atfi, A.; Watabe, K.; Lu, Z.; Mo, Y.Y. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 2016, 7, e2262. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Mateo, C.; Torres, B.; Gutiérrez, G.; Pintor-Toro, J.A. Downregulation of Lnc-Spry1 mediates TGF-β-induced epithelial-mesenchymal transition by transcriptional and posttranscriptional regulatory mechanisms. Cell Death Differ. 2017, 24, 785–797. [Google Scholar] [CrossRef]
- De Troyer, L.; Zhao, P.; Pastor, T.; Baietti, M.F.; Barra, J.; Vendramin, R.; Dok, R.; Lechat, B.; Najm, P.; Van Haver, D.; et al. Stress-induced lncRNA LASTR fosters cancer cell fitness by regulating the activity of the U4/U6 recycling factor SART3. Nucleic Acids Res. 2020, 48, 2502–2517. [Google Scholar] [CrossRef]
- van Dijk, M.; Visser, A.; Buabeng, K.M.L.; Poutsma, A.; van der Schors, R.C.; Oudejans, C.B.M. Mutations within the LINC-HELLP non-coding RNA differentially bind ribosomal and RNA splicing complexes and negatively affect trophoblast differentiation. Hum. Mol. Genet. 2015, 24, 5475–5485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhasnaoui, J.; Miano, V.; Ferrero, G.; Doria, E.; Leon, A.E.; Fabricio, A.S.C.; Annaratone, L.; Castellano, I.; Sapino, A.; De Bortoli, M. DSCAM-AS1-driven proliferation of breast cancer cells involves regulation of alternative exon splicing and 3′-end usage. Cancers 2020, 12, 1453. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell. Biol. 2016, 17, 205–211. [Google Scholar] [CrossRef]
- Xie, L.; Mao, M.; Xiong, K.; Jiang, B. Circular RNAs: A novel player in development and disease of the central nervous system. Front. Cell Neurosci. 2017, 11, 354. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, D.; Caponnetto, A.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; D’Angeli, F.; Morrone, A.; Caltabiano, R.; Barbagallo, G.M.; Ragusa, M.; et al. CircSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018, 19, 480. [Google Scholar] [CrossRef] [Green Version]
- Kelly, S.; Greenman, C.; Cook, P.R.; Papantonis, A. Exon Skipping Is Correlated with Exon Circularization. J. Mol. Biol. 2015, 427, 2414–2417. [Google Scholar] [CrossRef]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6 -methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pisignano, G.; Ladomery, M. Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message. Non-Coding RNA 2021, 7, 21. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010021
Pisignano G, Ladomery M. Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message. Non-Coding RNA. 2021; 7(1):21. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010021
Chicago/Turabian StylePisignano, Giuseppina, and Michael Ladomery. 2021. "Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message" Non-Coding RNA 7, no. 1: 21. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010021





