miR-24 Targets the Transmembrane Glycoprotein Neuropilin-1 in Human Brain Microvascular Endothelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Identification of miRNAs Targeting Neuropilin-1
2.3. Biological Validation of miR-24 as a Regulator of Neuropilin-1
2.4. Western Blot
2.5. Endothelial Permeability Assay
2.6. Statistical Analysis
3. Results
3.1. Identification of miR-24 as a Specific Modulator of Neuropilin-1
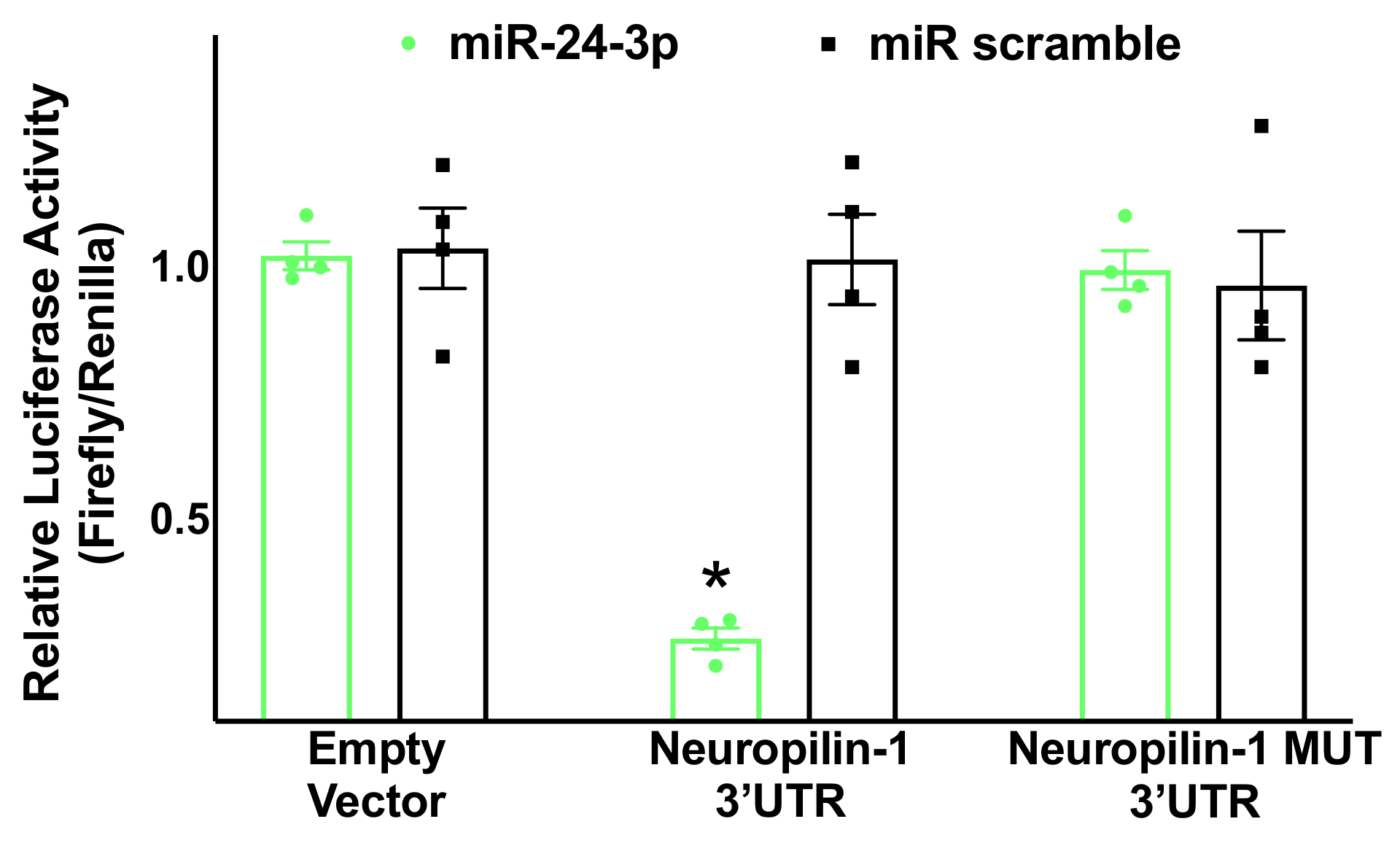
3.2. Neuropilin-1 Is a Molecular Target of miR-24
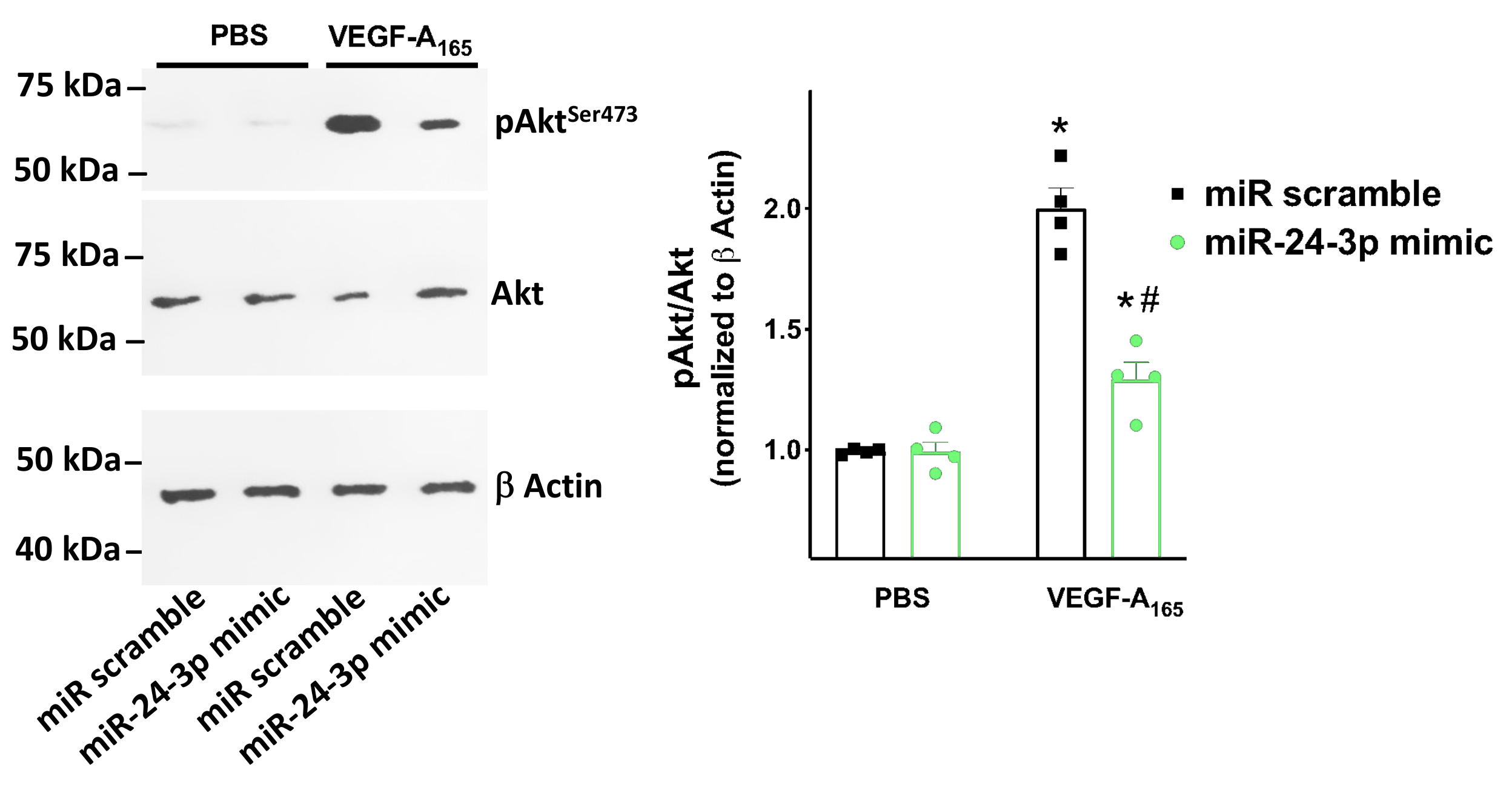
3.3. miR-24 Regulates Neuropilin-1 Transcription Levels in Human Endothelial Cells
3.4. miR-24 Regulates Neuropilin-1 Mediated Endothelial Permeability
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, Z.; Tessier-Lavigne, M. Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell 1997, 90, 739–751. [Google Scholar] [CrossRef] [Green Version]
- Kolodkin, A.L.; Levengood, D.V.; Rowe, E.G.; Tai, Y.T.; Giger, R.J.; Ginty, D.D. Neuropilin is a semaphorin III receptor. Cell 1997, 90, 753–762. [Google Scholar] [CrossRef] [Green Version]
- Soker, S.; Takashima, S.; Miao, H.Q.; Neufeld, G.; Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 1998, 92, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Oh, H.; Takagi, H.; Otani, A.; Koyama, S.; Kemmochi, S.; Uemura, A.; Honda, Y. Selective induction of neuropilin-1 by vascular endothelial growth factor (VEGF): A mechanism contributing to VEGF-induced angiogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 383–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellet-Many, C.; Frankel, P.; Jia, H.; Zachary, I. Neuropilins: Structure, function and role in disease. Biochem. J. 2008, 411, 211–226. [Google Scholar]
- Rossignol, M.; Gagnon, M.L.; Klagsbrun, M. Genomic organization of human neuropilin-1 and neuropilin-2 genes: Identification and distribution of splice variants and soluble isoforms. Genomics 2000, 70, 211–222. [Google Scholar] [CrossRef]
- Dumond, A.; Pages, G. Neuropilins, as relevant oncology target: Their role in the tumoral microenvironment. Front. Cell. Dev. Biol. 2020, 8, 662. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, Y.; Mangalam, A.K.; Guo, Y.; LaFrance-Corey, R.G.; Gamez, J.D.; Atanga, P.A.; Clarkson, B.D.; Zhang, Y.; Wang, E.; et al. Neuropilin-1 modulates interferon-gamma-stimulated signaling in brain microvascular endothelial cells. J. Cell. Sci. 2016, 129, 3911–3921. [Google Scholar] [CrossRef] [Green Version]
- Santulli, G. MicroRNAs distinctively regulate vascular smooth muscle and endothelial cells: Functional implications in angiogenesis, atherosclerosis, and in-stent restenosis. Adv. Exp. Med. Biol. 2015, 887, 53–77. [Google Scholar]
- Stavast, C.J.; Erkeland, S.J. The non-canonical aspects of MicroRNAs: Many roads to gene regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [Green Version]
- Van der Kwast, R.; Quax, P.H.A.; Nossent, A.Y. An emerging role for isomiRs and the microRNA epitranscriptome in Neovascularization. Cells 2019, 9, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santulli, G. Exosomal MicroRNA: The revolutionary endogenous innerspace nanotechnology. Sci. Transl. Med. 2018, 10, eaav9141. [Google Scholar] [CrossRef] [PubMed]
- Godlewski, J.; Lenart, J.; Salinska, E. MicroRNA in brain pathology: Neurodegeneration the other side of the brain cancer. Noncoding RNA 2019, 5, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wronska, A.; Kurkowska-Jastrzebska, I.; Santulli, G. Application of MicroRNAs in diagnosis and treatment of cardiovascular disease. Acta. Physiol. 2015, 213, 60–83. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.K.M.; Sorensen, A.E.; Joglekar, M.V.; Hardikar, A.A.; Dalgaard, L.T. Non-Coding RNA in pancreas and beta-cell development. Noncoding RNA 2018, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- Dama, E.; Melocchi, V.; Mazzarelli, F.; Colangelo, T.; Cuttano, R.; Di Candia, L.; Ferretti, G.M.; Taurchini, M.; Graziano, P.; Bianchi, F. Non-Coding RNAs as Prognostic Biomarkers: A miRNA signature specific for aggressive early-stage lung adenocarcinomas. Noncoding RNA 2020, 6, 48. [Google Scholar] [CrossRef]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating MicroRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef]
- Fay, E.J.; Langlois, R.A. MicroRNA-attenuated virus vaccines. Noncoding RNA 2018, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Bar, C.; Chatterjee, S.; Falcao Pires, I.; Rodrigues, P.; Sluijter, J.P.G.; Boon, R.A.; Nevado, R.M.; Andres, V.; Sansonetti, M.; de Windt, L.; et al. Non-coding RNAs: Update on mechanisms and therapeutic targets from the ESC Working Groups of Myocardial Function and Cellular Biology of the Heart. Cardiovasc. Res. 2020, 116, 1805–1819. [Google Scholar] [CrossRef]
- Slota, J.A.; Booth, S.A. MicroRNAs in neuroinflammation: Implications in disease pathogenesis, biomarker discovery and therapeutic applications. Noncoding RNA 2019, 5, 35. [Google Scholar] [CrossRef] [Green Version]
- Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V.; Bansal, P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016, 7, 68–74. [Google Scholar] [PubMed]
- Santulli, G. MicroRNA: From Molecular Biology to Clinical Practice; Santulli, G., Ed.; Springer Nature: New York, NY, USA, 2016. [Google Scholar]
- Yang, Y.; Liu, Y.; Li, Y.; Chen, Z.; Xiong, Y.; Zhou, T.; Tao, W.; Xu, F.; Yang, H.; Yla-Herttuala, S.; et al. MicroRNA-15b targets VEGF and inhibits angiogenesis in proliferative diabetic retinopathy. J. Clin. Endocrinol. Metab. 2020, 105, 3404–3415. [Google Scholar] [CrossRef] [PubMed]
- Widlansky, M.E.; Jensen, D.M.; Wang, J.; Liu, Y.; Geurts, A.M.; Kriegel, A.J.; Liu, P.; Ying, R.; Zhang, G.; Casati, M.; et al. MiR-29 contributes to normal endothelial function and can restore it in cardiometabolic disorders. EMBO. Mol. Med. 2018, 10, e8046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santulli, G. MicroRNAs and Endothelial (Dys) Function. J. Cell. Physiol. 2016, 231, 1638–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific MicroRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008, 15, 261–271. [Google Scholar] [CrossRef] [Green Version]
- Rafehi, H.; El-Osta, A. HDAC inhibition in vascular endothelial cells regulates the expression of ncRNAs. Noncoding RNA 2016, 2, 4. [Google Scholar] [CrossRef]
- Santulli, G.; Wronska, A.; Uryu, K.; Diacovo, T.G.; Gao, M.; Marx, S.O.; Kitajewski, J.; Chilton, J.M.; Akat, K.M.; Tuschl, T.; et al. A selective microRNA-based strategy inhibits restenosis while preserving endothelial function. J. Clin. Investig. 2014, 124, 4102–4114. [Google Scholar] [CrossRef]
- Eigenmann, D.E.; Xue, G.; Kim, K.S.; Moses, A.V.; Hamburger, M.; Oufir, M. Comparative study of four immortalized human brain capillary endothelial cell lines, hCMEC/D3, hBMEC, TY10, and BB19, and optimization of culture conditions, for an in vitro blood-brain barrier model for drug permeability studies. Fluids. Barriers. CNS 2013, 10, 33. [Google Scholar] [CrossRef] [Green Version]
- Matarese, A.; Gambardella, J.; Lombardi, A.; Wang, X.; Santulli, G. MiR-7 Regulates GLP-1-Mediated Insulin Release by Targeting beta-Arrestin 1. Cells 2020, 9, 1621. [Google Scholar] [CrossRef]
- Wang, X.; Morelli, M.B.; Matarese, A.; Sardu, C.; Santulli, G. Cardiomyocyte-derived exosomal microRNA-92a mediates post-ischemic myofibroblast activation both in vitro and ex vivo. ESC. Heart. Fail. 2020, 7, 284–288. [Google Scholar] [CrossRef] [Green Version]
- Matarese, A.; Gambardella, J.; Sardu, C.; Santulli, G. MiR-98 regulates TMPRSS2 expression in human endothelial cells: Key implications for COVID-19. Biomedicines 2020, 8, 642. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.B.; Shu, J.; Sardu, C.; Matarese, A.; Santulli, G. Cardiosomal microRNAs are essential in post-infarction myofibroblast phenoconversion. Int. J. Mol. Sci. 2019, 21, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gambardella, J.; Sorriento, D.; Bova, M.; Rusciano, M.; Loffredo, S.; Wang, X.; Petraroli, A.; Carucci, L.; Mormile, I.; Oliveti, M.; et al. Role of endothelial G Protein-coupled receptor kinase 2 in angioedema. Hypertension 2020, 76, 1625–1636. [Google Scholar] [CrossRef]
- Becker, P.M.; Waltenberger, J.; Yachechko, R.; Mirzapoiazova, T.; Sham, J.S.; Lee, C.G.; Elias, J.A.; Verin, A.D. Neuropilin-1 regulates vascular endothelial growth factor-mediated endothelial permeability. Circ. Res. 2005, 96, 1257–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, L.; Prahst, C.; Ruckdeschel, T.; Savant, S.; Westrom, S.; Fantin, A.; Riedel, M.; Heroult, M.; Ruhrberg, C.; Augustin, H.G. Neuropilin-1 mediates vascular permeability independently of vascular endothelial growth factor receptor-2 activation. Sci. Signal. 2016, 9, 42. [Google Scholar] [CrossRef] [Green Version]
- Fantin, A.; Lampropoulou, A.; Senatore, V.; Brash, J.T.; Prahst, C.; Lange, C.A.; Liyanage, S.E.; Raimondi, C.; Bainbridge, J.W.; Augustin, H.G.; et al. VEGF165-induced vascular permeability requires NRP1 for ABL-mediated SRC family kinase activation. J. Exp. Med. 2017, 214, 1049–1064. [Google Scholar] [CrossRef]
- Tymecka, D.; Lipinski, P.F.J.; Fedorczyk, B.; Puszko, A.; Wilenska, B.; Perret, G.Y.; Misicka, A. Structure-activity relationship study of tetrapeptide inhibitors of the Vascular Endothelial Growth Factor A binding to Neuropilin-1. Peptides 2017, 94, 25–32. [Google Scholar] [CrossRef]
- Narazaki, M.; Tosato, G. Ligand-induced internalization selects use of common receptor neuropilin-1 by VEGF165 and semaphorin3A. Blood 2006, 107, 3892–3901. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Zhou, Z.; Fei, L.; Sun, H.; Wang, R.; Chen, Y.; Chen, H.; Wang, J.; Tang, H.; Ge, W.; et al. Construction of a human cell landscape at single-cell level. Nature 2020, 581, 303–309. [Google Scholar] [CrossRef]
- Hsieh, S.H.; Ying, N.W.; Wu, M.H.; Chiang, W.F.; Hsu, C.L.; Wong, T.Y.; Jin, Y.T.; Hong, T.M.; Chen, Y.L. Galectin-1, a novel ligand of neuropilin-1, activates VEGFR-2 signaling and modulates the migration of vascular endothelial cells. Oncogene 2008, 27, 3746–3753. [Google Scholar] [CrossRef] [Green Version]
- Fiedler, J.; Jazbutyte, V.; Kirchmaier, B.C.; Gupta, S.K.; Lorenzen, J.; Hartmann, D.; Galuppo, P.; Kneitz, S.; Pena, J.T.; Sohn-Lee, C.; et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 2011, 124, 720–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Ouyang, L.; He, L.; Qu, Y.; Han, Y.; Duan, D. Inhibition of exosomal miR-24-3p in diabetes restores angiogenesis and facilitates wound repair via targeting PIK3R3. J. Cell. Mol. Med. 2020, 24, 13789–13803. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, Y.; Liu, G.; Qi, X.; Cao, X. MicroRNA-24 inhibits the proliferation and migration of endothelial cells in patients with atherosclerosis by targeting importin-alpha3 and regulating inflammatory responses. Exp. Ther. Med. 2018, 15, 338–344. [Google Scholar]
- Davies, J.; Randeva, H.S.; Chatha, K.; Hall, M.; Spandidos, D.A.; Karteris, E.; Kyrou, I. Neuropilin1 as a new potential SARSCoV2 infection mediator implicated in the neurologic features and central nervous system involvement of COVID19. Mol. Med. Rep. 2020, 22, 4221–4226. [Google Scholar] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar]
- Santulli, G.; Morelli, M.; Gambardella, J. Is Endothelial Dysfunction the Concealed Cornerstone of COVID-19? BMJ. 2020, 368, m1091. [Google Scholar] [CrossRef] [Green Version]
- Sardu, C.; Gambardella, J.; Morelli, M.B.; Wang, X.; Marfella, R.; Santulli, G. Hypertension, Thrombosis, Kidney Failure, and Diabetes: Is COVID-19 an Endothelial Disease? A Comprehensive Evaluation of Clinical and Basic Evidence. J. Clin. Med. 2020, 9, 1417. [Google Scholar] [CrossRef]
- Gambardella, J.; Santulli, G. What is linking COVID-19 and endothelial dysfunction? Updates on nanomedicine and bioengineering from the 2020 AHA Scientific Sessions. Eur. Heart. J. Cardiovasc. Pharmacother. 2020, 30, pvaa145. [Google Scholar] [CrossRef]
- Kaur, S.; Tripathi, D.M.; Yadav, A. The Enigma of Endothelium in COVID-19. Front. Physiol. 2020, 11, 989. [Google Scholar] [CrossRef]
- Evans, P.C.; Rainger, G.E.; Mason, J.C.; Guzik, T.J.; Osto, E.; Stamataki, Z.; Neil, D.; Hoefer, I.E.; Fragiadaki, M.; Waltenberger, J.; et al. Endothelial dysfunction in COVID-19: A position paper of the ESC Working Group for Atherosclerosis and Vascular Biology, and the ESC Council of Basic Cardiovascular Science. Cardiovasc. Res. 2020, 116, 2177–2184. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Luscher, T. COVID-19 is, in the end, an endothelial disease. Eur. Heart J. 2020, 41, 3038–3044. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.E.; Curley, G.F.; Lavin, M.; Fogarty, H.; Karampini, E.; McEvoy, N.L.; Clarke, J.; Boylan, M.; Alalqam, R.; Worrall, A.P.; et al. Von Willebrand factor propeptide in severe coronavirus disease 2019 (COVID-19): Evidence of acute and sustained endothelial cell activation. Br. J. Haematol. 2020. [Google Scholar] [CrossRef]
- Fraser, D.D.; Patterson, E.K.; Slessarev, M.; Gill, S.E.; Martin, C.; Daley, M.; Miller, M.R.; Patel, M.A.; Dos Santos, C.C.; Bosma, K.J.; et al. Endothelial Injury and Glycocalyx Degradation in Critically Ill Coronavirus Disease 2019 Patients: Implications for Microvascular Platelet Aggregation. Crit. Care. Explor. 2020, 2, e0194. [Google Scholar] [CrossRef] [PubMed]
- Loo, J.; Spittle, D.A.; Newnham, M. COVID-19, immunothrombosis and venous thromboembolism: Biological mechanisms. Thorax 2021. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.A.; Frishman, W.H. Thrombotic Complications of COVID-19 Infection: A Review. Cardiol. Rev. 2021, 29, 43–47. [Google Scholar] [CrossRef]
- Sardu, C.; Gambardella, J.; Morelli, M.; Wang, X.; Marfella, R.; Santulli, G. Is COVID-19 an Endothelial Disease? Clinical and Basic Evidence. Preprints 2020, 2020040204. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef]
- Fox, S.E.; Li, G.; Akmatbekov, A.; Harbert, J.L.; Lameira, F.S.; Brown, J.Q.; Vander Heide, R.S. Unexpected Features of Cardiac Pathology in COVID-19 Infection. Circulation 2020, 142, 1123–1125. [Google Scholar] [CrossRef]
- Stahl, K.; Brasen, J.H.; Hoeper, M.M.; David, S. Direct evidence of SARS-CoV-2 in gut endothelium. Intensive Care Med. 2020, 46, 2081–2082. [Google Scholar] [CrossRef] [PubMed]
- Bosmuller, H.; Traxler, S.; Bitzer, M.; Haberle, H.; Raiser, W.; Nann, D.; Frauenfeld, L.; Vogelsberg, A.; Klingel, K.; Fend, F. The evolution of pulmonary pathology in fatal COVID-19 disease: An autopsy study with clinical correlation. Virchows Arch. 2020, 477, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Ilonzo, N.; Kumar, S.; Borazan, N.; Hansen, T.; Rao, A.; Lantis, J.; Faries, P.; Ting, W. Endotheliitis in COVID-19-positive patients after extremity amputation for acute thrombotic events. Ann. Vasc. Surg. 2020, 30. [Google Scholar] [CrossRef]
- Buzhdygan, T.P.; DeOre, B.J.; Baldwin-Leclair, A.; Bullock, T.A.; McGary, H.M.; Khan, J.A.; Razmpour, R.; Hale, J.F.; Galie, P.A.; Potula, R.; et al. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood-brain barrier. Neurobiol. Dis. 2020, 146, 105131. [Google Scholar] [CrossRef] [PubMed]
- Zubair, A.S.; McAlpine, L.S.; Gardin, T.; Farhadian, S.; Kuruvilla, D.E.; Spudich, S. Neuropathogenesis and Neurologic Manifestations of the Coronaviruses in the Age of Coronavirus Disease 2019: A Review. JAMA Neurol. 2020, 77, 1018–1027. [Google Scholar] [CrossRef]
- Desai, I.; Manchanda, R.; Kumar, N.; Tiwari, A.; Kumar, M. Neurological manifestations of coronavirus disease 2019: Exploring past to understand present. Neurol. Sci. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Tandon, M.; Kataria, S.; Patel, J.; Mehta, T.R.; Daimee, M.; Patel, V.; Prasad, A.; Chowdhary, A.A.; Jaiswal, S.; Sriwastava, S. A Comprehensive Systematic Review of CSF analysis that defines Neurological Manifestations of COVID-19. Int. J. Infect. Dis. 2021, 9. [Google Scholar] [CrossRef]
- Paniz-Mondolfi, A.; Bryce, C.; Grimes, Z.; Gordon, R.E.; Reidy, J.; Lednicky, J.; Sordillo, E.M.; Fowkes, M. Central nervous system involvement by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). J. Med. Virol. 2020, 92, 699–702. [Google Scholar] [CrossRef] [Green Version]
- Rhea, E.M.; Logsdon, A.F.; Hansen, K.M.; Williams, L.M.; Reed, M.J.; Baumann, K.K.; Holden, S.J.; Raber, J.; Banks, W.A.; Erickson, M.A. The S1 protein of SARS-CoV-2 crosses the blood-brain barrier in mice. Nat. Neurosci. 2020. [Google Scholar] [CrossRef]
- Mayi, B.S.; Leibowitz, J.A.; Woods, A.T.; Ammon, K.A.; Liu, A.E.; Raja, A. The role of Neuropilin-1 in COVID-19. PLoS Pathog. 2021, 17, e1009153. [Google Scholar] [CrossRef]
- Teesalu, T.; Sugahara, K.N.; Kotamraju, V.R.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2009, 106, 16157–16162. [Google Scholar] [CrossRef] [Green Version]
- Ray, P.R.; Wangzhou, A.; Ghneim, N.; Yousuf, M.S.; Paige, C.; Tavares-Ferreira, D.; Mwirigi, J.M.; Shiers, S.; Sankaranarayanan, I.; McFarland, A.J.; et al. A pharmacological interactome between COVID-19 patient samples and human sensory neurons reveals potential drivers of neurogenic pulmonary dysfunction. Brain Behav. Immun. 2020, 89, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.X.; Tang, X.J.; Shi, Q.L.; Li, Q.; Deng, H.J.; Yuan, J.; Hu, J.L.; Xu, W.; Zhang, Y.; Lv, F.J.; et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020, 26, 1200–1204. [Google Scholar] [CrossRef] [PubMed]
- Moutal, A.; Martin, L.F.; Boinon, L.; Gomez, K.; Ran, D.; Zhou, Y.; Stratton, H.J.; Cai, S.; Luo, S.; Gonzalez, K.B.; et al. SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain 2021, 162, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Karakiulakis, G.; Roth, M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020, 8, e21. [Google Scholar] [PubMed]
- Rubino, F.; Amiel, S.A.; Zimmet, P.; Alberti, G.; Bornstein, S.; Eckel, R.H.; Mingrone, G.; Boehm, B.; Cooper, M.E.; Chai, Z.; et al. New-Onset Diabetes in Covid-19. N. Engl. J. Med. 2020, 383, 789–790. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.C.; Wu, H.; Kirita, Y.; Uchimura, K.; Ledru, N.; Rennke, H.G.; Welling, P.A.; Waikar, S.S.; Humphreys, B.D. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl. Acad. Sci. USA 2019, 116, 19619–19625. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Cheng, J.; Wang, D.; Hu, X.; Xie, Y.; Stitham, J.; Atteya, G.; Du, J.; Tang, W.H.; Lee, S.H.; et al. Hyperglycemia repression of miR-24 coordinately upregulates endothelial cell expression and secretion of von Willebrand factor. Blood 2015, 125, 3377–3387. [Google Scholar] [CrossRef]
- Flumignan, R.L.; Tinôco, J.D.S.; Pascoal, P.I.; Areias, L.L.; Cossi, M.S.; Fernandes, M.I.; Costa, I.K.; Souza, L.; Matar, C.F.; Tendal, B.; et al. Prophylactic anticoagulants for people hospitalised with COVID-19. Cochrane Database Syst. Rev. 2020, 10. [Google Scholar] [CrossRef]
- McFadyen, J.D.; Stevens, H.; Peter, K. The Emerging Threat of (Micro) Thrombosis in COVID-19 and Its Therapeutic Implications. Circ. Res. 2020, 127, 571–587. [Google Scholar] [CrossRef]
- Vinayagam, S.; Sattu, K. SARS-CoV-2 and coagulation disorders in different organs. Life. Sci. 2020, 260, 118431. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, B.; Khaled, Y.S.; Ammori, B.J.; Elkord, E. Neuropilin 1: Function and therapeutic potential in cancer. Cancer Immunol. Immunother. 2014, 63, 81–99. [Google Scholar] [CrossRef] [PubMed]
- Thepmankorn, P.; Bach, J.; Lasfar, A.; Zhao, X.; Souayah, S.; Chong, Z.Z.; Souayah, N. Cytokine storm induced by SARS-CoV-2 infection: The spectrum of its neurological manifestations. Cytokine 2020, 138, 155404. [Google Scholar] [CrossRef] [PubMed]
- Nigrovic, P.A. COVID-19 cytokine storm: What is in a name? Ann. Rheum. Dis. 2021, 80, 3–5. [Google Scholar] [CrossRef]
- Brunetti, L.; Diawara, O.; Tsai, A.; Firestein, B.L.; Nahass, R.G.; Poiani, G.; Schlesinger, N. Colchicine to Weather the Cytokine Storm in Hospitalized Patients with COVID-19. J. Clin. Med. 2020, 9, 2961. [Google Scholar] [CrossRef]
- Connor, K.L.; Teenan, O.; Cairns, C.; Banwell, V.; Thomas, R.A.; Rodor, J.; Finnie, S.; Pius, R.; Tannahill, G.M.; Sahni, V.; et al. Identifying cell-enriched miRNAs in kidney injury and repair. JCI Insight 2020, 5. [Google Scholar] [CrossRef]
- Dai, D.; Huang, W.; Lu, Q.; Chen, H.; Liu, J.; Hong, B. miR24 regulates angiogenesis in gliomas. Mol. Med. Rep. 2018, 18, 358–368. [Google Scholar]
- Zhang, J.; Liu, L.; Xue, Y.; Ma, Y.; Liu, X.; Li, Z.; Li, Z.; Liu, Y. Endothelial Monocyte-Activating Polypeptide-II Induces BNIP3-Mediated Mitophagy to Enhance Temozolomide Cytotoxicity of Glioma Stem Cells via Down-Regulating MiR-24-3p. Front. Mol. Neurosci. 2018, 11, 92. [Google Scholar] [CrossRef]
- Li, H.T.; Wang, J.; Li, S.F.; Cheng, L.; Tang, W.Z.; Feng, Y.G. Upregulation of microRNA24 causes vasospasm following subarachnoid hemorrhage by suppressing the expression of endothelial nitric oxide synthase. Mol. Med. Rep. 2018, 18, 1181–1187. [Google Scholar]



| Gene | Primer | Sequence (5′–3′) | Amplicon (bp) |
|---|---|---|---|
| Neuropilin-1 | Forward | CCA CAG TGG AAC AGG TGA TG | 114 |
| Reverse | ACA CAC ACA GGC GTT AGC TG | ||
| GAPDH | Forward | GGC TCC CTT GGG TAT ATG GT | 94 |
| Reverse | TTG ATT TTG GAG GGA TCT CG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mone, P.; Gambardella, J.; Wang, X.; Jankauskas, S.S.; Matarese, A.; Santulli, G. miR-24 Targets the Transmembrane Glycoprotein Neuropilin-1 in Human Brain Microvascular Endothelial Cells. Non-Coding RNA 2021, 7, 9. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010009
Mone P, Gambardella J, Wang X, Jankauskas SS, Matarese A, Santulli G. miR-24 Targets the Transmembrane Glycoprotein Neuropilin-1 in Human Brain Microvascular Endothelial Cells. Non-Coding RNA. 2021; 7(1):9. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010009
Chicago/Turabian StyleMone, Pasquale, Jessica Gambardella, Xujun Wang, Stanislovas S. Jankauskas, Alessandro Matarese, and Gaetano Santulli. 2021. "miR-24 Targets the Transmembrane Glycoprotein Neuropilin-1 in Human Brain Microvascular Endothelial Cells" Non-Coding RNA 7, no. 1: 9. https://0-doi-org.brum.beds.ac.uk/10.3390/ncrna7010009







